For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KPVDVVEPVSEPTTPKDMRKICAAGAVGTGIELYDFLIFGLAAGLVFPKLFFPGSDPLTGALLSFMALGA GFVSRPLGGAVFGHFGDRVSRKTMLVVSLVATGLCTMGIGALPTYASIGTLAPILLVTLRVLQGFFMGGE QAGAFIVVTEHAPTGRRAFFGASVTAGSPLGSILGIGVFNIVALATGAAFVEWGWRIPFLASAVLVAVGL YVRLSIGESPVFRRIAAQRAAVRAVPLWGALKGAPHLLIGGVLVNLGFNLFIFIINSFTSSYGTQQLGIE RDDILLSGLWGSFGMLVFVFVAGGLADRFGLVRVMLCGAIFQVLWAYPYFWLLDTRGIPALYVAVVVAYI GLSFVFGPMAAFYVSLFRPEIRYSGVAVSYNLGAVLGGGLSPAIATALVQRYDASTGISAYIALGGVVSA LGLLLTARQVKAARDPADI*
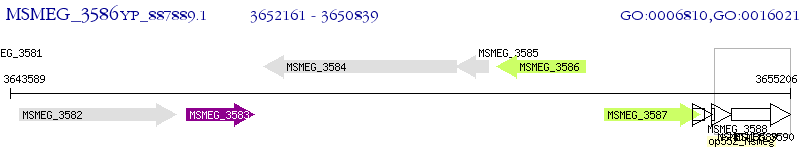
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3586 | - | - | 100% (440) | major facilitator family protein transporter |
| M. smegmatis MC2 155 | MSMEG_4122 | - | 2e-88 | 42.24% (419) | major facilitator family protein transporter |
| M. smegmatis MC2 155 | MSMEG_3386 | - | 2e-85 | 39.24% (423) | shikimate transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1232 | - | 2e-75 | 39.56% (412) | integral membrane transport protein |
| M. gilvum PYR-GCK | Mflv_4524 | - | 1e-89 | 40.82% (414) | major facilitator transporter |
| M. tuberculosis H37Rv | Rv1200 | - | 2e-75 | 39.56% (412) | integral membrane transport protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3416 | - | 1e-69 | 37.72% (403) | membrane transporter |
| M. marinum M | MMAR_4238 | - | 4e-71 | 37.83% (423) | integral membrane transport protein |
| M. avium 104 | MAV_1344 | - | 3e-75 | 40.18% (438) | major facilitator family protein transporter |
| M. thermoresistible (build 8) | TH_3714 | - | 4e-86 | 43.06% (418) | PUTATIVE major facilitator superfamily MFS_1 |
| M. ulcerans Agy99 | MUL_0949 | - | 1e-70 | 37.59% (423) | integral membrane transport protein |
| M. vanbaalenii PYR-1 | Mvan_2899 | - | 1e-83 | 40.20% (408) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4524|M.gilvum_PYR-GCK -------MTTATPSDPATLRRVALSSLLGTAIEYYDFLVYGTMSALVFGR
Mvan_2899|M.vanbaalenii_PYR-1 -------MNEASVADSALLRRVALSSLLGTAIEYYDFLVYGTMSALVFGL
TH_3714|M.thermoresistible__bu -----MSAEAATPTSRVTPRRVAVASFIGTTVEFYDFLIYGTAAALVFPK
MSMEG_3586|M.smegmatis_MC2_155 MKPVDVVEPVSEPTTPKDMRKICAAGAVGTGIELYDFLIFGLAAGLVFPK
Mb1232|M.bovis_AF2122/97 ------------------MKRVALACLVGSAIEFYDFLIYGTAAALVFPT
Rv1200|M.tuberculosis_H37Rv ------------------MKRVALACLVGSAIEFYDFLIYGTAAALVFPT
MMAR_4238|M.marinum_M ------------------MSRVALACVVGSTVEYYDFCIYGTAAALVFPA
MUL_0949|M.ulcerans_Agy99 ------------------MSRVALACVVGSTVEYYDFCIYGTAAALVFPA
MAV_1344|M.avium_104 ----MGEPSARATTPGTPMRRIAAACLVGSAIEFYDFLIYGTAAALVFPA
MAB_3416|M.abscessus_ATCC_1997 -MTSSPAEPRQMADGQVSPRRVFMASAVGSAIEFYDFYIYGTAAALVFPA
:: : :*: :* *** ::* :.***
Mflv_4524|M.gilvum_PYR-GCK VFFPDSDPAVATIAAFGTLAAGYVARPVGGILFGHFGDRLGRKRMLVLTM
Mvan_2899|M.vanbaalenii_PYR-1 VFFPDSDPAVATIAAFGTLAAGYVARPVGGIVFGHFGDRIGRKSMLVLTM
TH_3714|M.thermoresistible__bu LFFPTASPASGVLLSFATFGVGFFARPLGGIVFGHFGDRLGRKRMLVYSL
MSMEG_3586|M.smegmatis_MC2_155 LFFPGSDPLTGALLSFMALGAGFVSRPLGGAVFGHFGDRVSRKTMLVVSL
Mb1232|M.bovis_AF2122/97 VFFPHLDPTVAAVASMGTFAVAFLSRPFGAAVFGYFGDRLGRKKTLVATL
Rv1200|M.tuberculosis_H37Rv VFFPHLDPTVAAVASMGTFAVAFLSRPFGAAVFGYFGDRLGRKKTLVATL
MMAR_4238|M.marinum_M VFYPGLSPALATIASMGTFATAFLSRPLGAAVFGHFGDRLGRKKTLVATL
MUL_0949|M.ulcerans_Agy99 VFYPGLSPALATIASMGTFATAFLSRPLGAAVFGHFGDRLGRKKTLVATL
MAV_1344|M.avium_104 VFFPRLGPTVATIASMATFATAFLSRPLGAAVFGYFGDRLGRKKTLVATL
MAB_3416|M.abscessus_ATCC_1997 VFFPNLSHSLALFASIATFSTAFLARPIGAALFGHYGDRFSRKGTLVVTL
:*:* . . . :: ::...:.:**.*. :**::***..** ** ::
Mflv_4524|M.gilvum_PYR-GCK GLMGAASFAIGLLPTFAAVGVTAPLLLVALRVIQGIAIGGEWGGATLMVT
Mvan_2899|M.vanbaalenii_PYR-1 TLMGTASFLIGLLPSFAAVGVAAPILLVALRVIQGVAIGGEWGGATLMVT
TH_3714|M.thermoresistible__bu VGMGVATVLIGLLPTYAQIGMAAPALLTLLRLAQGFAMGGEWGGATLMAV
MSMEG_3586|M.smegmatis_MC2_155 VATGLCTMGIGALPTYASIGTLAPILLVTLRVLQGFFMGGEQAGAFIVVT
Mb1232|M.bovis_AF2122/97 LIMGLATVTVGLVPTTVAIGAAAPLILTTMRLLQGFAVGGEWAGSALLSA
Rv1200|M.tuberculosis_H37Rv LIMGLATVTVGLVPTTVAIGAAAPLILTTMRLLQGFAVGGEWAGSALLSA
MMAR_4238|M.marinum_M LIMALSTVTVGVVPSTATIGVSAPLILVALRLLQGFAVGGEWAGSALLSI
MUL_0949|M.ulcerans_Agy99 LIMALSTVTVGVVPSTATIGVSAPLILVALRLLQGFAVGGEWAGSALLSI
MAV_1344|M.avium_104 LIMGASTVSVGLVPSTASIGIAAPLLLTVLRLLQGFAVGAEWAGSVLLSA
MAB_3416|M.abscessus_ATCC_1997 ITMGLATMAVGLVPGAATIGAAAPTAIVILRLVQGLAVGGEWSGAALLSA
. .:. :* :* . :* ** :. :*: **. :*.* .*: ::
Mflv_4524|M.gilvum_PYR-GCK EHAEPHRRGFWNGVMQMGSPIGSLLSVTVVTAITLLPDE---AFVSWGWR
Mvan_2899|M.vanbaalenii_PYR-1 EHAEAHRRGFWNGVMQMGSPIGSLLSVAVVTVITLLPDE---SFLSWGWR
TH_3714|M.thermoresistible__bu EHASADRKGFYGAFPQMGAPAGTATATLAFFAVSRLPDG---QFLSWGWR
MSMEG_3586|M.smegmatis_MC2_155 EHAPTGRRAFFGASVTAGSPLGSILGIGVFNIVALATGA---AFVEWGWR
Mb1232|M.bovis_AF2122/97 EYAPASKRGWYGMFTVVGGGIALVLTSLTFLGVNYTIGESSPTFMQWGWR
Rv1200|M.tuberculosis_H37Rv EYAPASKRGWYGMFTVVGGGIALVLTSLTFLGVNYTIGESSPTFMQWGWR
MMAR_4238|M.marinum_M ESAPAHKRGYYGMFTVLGGGVALVVSGLTFLGVNYTIGESSSAFLQWGWR
MUL_0949|M.ulcerans_Agy99 ESAPAHKRGYYGMFTVLGGGVALVVSGLTFLGVNYTIGESSSALLQWGWR
MAV_1344|M.avium_104 EYAPTDRRGWYGMFTLLGGGTAGILASLTFLAVNLTMGEHSPAFMHWGWR
MAB_3416|M.abscessus_ATCC_1997 EYSPSQNRGRAATAVPIGTSTGLLLSSLVFLIVSLTIGEDSPAFLSWGWR
* : . .:. * . .. : . :: ****
Mflv_4524|M.gilvum_PYR-GCK VPFLLSVVLLAIGVYVRVSITESPVFEQ----AKPKVVE-SRRIPLIEVL
Mvan_2899|M.vanbaalenii_PYR-1 IPFLVSLLLLAVGIYVRLSVTESPVFEA----ARKEAAGNAPRVPFVEIL
TH_3714|M.thermoresistible__bu LPFLLSAVLIGIGLAVRLTLTESPDFAA----VQARSAV--HRMPIAEAF
MSMEG_3586|M.smegmatis_MC2_155 IPFLASAVLVAVGLYVRLSIGESPVFRR----IAAQRAA-VRAVPLWGAL
Mb1232|M.bovis_AF2122/97 IPFLVSAALIAVALYVRFNIDETPVFAR--ERADEKTRLGPAETPIAQVL
Rv1200|M.tuberculosis_H37Rv IPFLVSAALIAVALYVRFNIDETPVFAR--ERADEKTRLGPAETPIAQVL
MMAR_4238|M.marinum_M VPFLLSALLIAFALYVRLEINETPVFARAMAQADDQESTAAARAPLAAVL
MUL_0949|M.ulcerans_Agy99 VPFLLSALLIAFALYVRLEINETPVFARAMAQADDQESTAAARAPLAAVL
MAV_1344|M.avium_104 VPFLISSGLIGIALYVRLNIDETPIFVE------EKARHLVPKAPLTELL
MAB_3416|M.abscessus_ATCC_1997 VPFIASGLLVAVGLYIRLQIAETPQFVE------AKAHHQLVKSPLRHVI
:**: * *:...: :*. : *:* * . *: :
Mflv_4524|M.gilvum_PYR-GCK RRPR-NLVLACAVGIGPFALTALISTHMISYATSIG-YHRTDVMTSLVFT
Mvan_2899|M.vanbaalenii_PYR-1 RRPR-TLVLACAVGIGPFALTALISTYMISYATAIG-YHRTDVMTALIFT
TH_3714|M.thermoresistible__bu RRHRRQILLVAGAYLSQGVFAYICVAYLVSYGTSVAGIDRTATLFGVSVA
MSMEG_3586|M.smegmatis_MC2_155 KGAPHLLIGGVLVNLGFNLFIFIINSFTSSYGTQQLGIERDDILLSGLWG
Mb1232|M.bovis_AF2122/97 RRQRREIVLAAGSAVCCFGFVYLASTYLASYAQTRLGYSRGSILFDSVLG
Rv1200|M.tuberculosis_H37Rv RRQRREIVLAAGSAVCCFGFVYLASTYLASYAQTRLGYSRGSILFDSVLG
MMAR_4238|M.marinum_M CRQRREIVLAAGAMLAAFGCLYLASTFLPSYAQLHLGYSRNLVLLIGVLG
MUL_0949|M.ulcerans_Agy99 CRQRREIVLAAGAMLAAFGCLYLASTFLPSYAQLHLGYSRNLVLLIGVLG
MAV_1344|M.avium_104 RLQRREIILVAGSFVGGMGFIYLGNTFLVMYAHNHLGYSRSFIWGIGALG
MAB_3416|M.abscessus_ATCC_1997 TESPRTLVLTSGTMLIIFTLSFMTNTYFPGFARSELHFSRAAILAAGALG
:: : : :. :. *
Mflv_4524|M.gilvum_PYR-GCK SLSALIAIPVFSALSDRVGRRPVMLVGGVAIICYAWPFYWLVDM-RAMAW
Mvan_2899|M.vanbaalenii_PYR-1 SLTGLIAIPFFSALSDHVGRRLVMVAGAIGIIGYAWPFYALVDM-RSQAY
TH_3714|M.thermoresistible__bu AVLAVVMYPLFGSLSDTFGRKPLFLAGVIAMGVSIGPAFALIDTGRPALF
MSMEG_3586|M.smegmatis_MC2_155 SFGMLVFVFVAGGLADRFGLVRVMLCGAIFQVLWAYPYFWLLDT-RGIPA
Mb1232|M.bovis_AF2122/97 GLLCIVFTALSSALCDQLGRRRVLLAGWAVALPWSLLVMPLIDS-GSPSL
Rv1200|M.tuberculosis_H37Rv GLLCIVFTALSSALCDQLGRRRVLLAGWAVALPWSLLVMPLIDS-GSPSL
MMAR_4238|M.marinum_M GLVCIGFVALSAILCDRFGRRRVMLVGWAVGLFWSPLVMPLIGS-GDTVG
MUL_0949|M.ulcerans_Agy99 GLVCIGFVALSAILCDRFGRRRVMLVGWAVGLFWSPLVMPLIGS-GDTVG
MAV_1344|M.avium_104 GLTSMACVACSAWISDRVGRRRVMLWGLVACLPWAFVVIPLIDT-GRPVC
MAB_3416|M.abscessus_ATCC_1997 AVVLMVSATAGGILSDRLGRRTVILSSMTLTLPWTLLVTPLLTS-GPFWA
.. : . :.* .* ::: . *:
Mflv_4524|M.gilvum_PYR-GCK LVVAMMVAQIVQSLMYAPLGALYSEMFGTTVRYTGASMGYQLAALVGAGF
Mvan_2899|M.vanbaalenii_PYR-1 LTAAMVIAQLIQSMMYAPLGALFSEMFGTTVRYTGASMGYQLAALLGAGF
TH_3714|M.thermoresistible__bu LVALVLVFGLAMAPAAGVTGALFAMTFDADVRYSGVSIGYTLSQVLGSAF
MSMEG_3586|M.smegmatis_MC2_155 LYVAVVVAYIGLSFVFGPMAAFYVSLFRPEIRYSGVAVSYNLGAVLGGGL
Mb1232|M.bovis_AF2122/97 FAVAVVGMYAIGGFGFGPTASFIPELFATSYRYTGSALAANLAGVAGGAL
Rv1200|M.tuberculosis_H37Rv FAVAVVGMYAIGGFGFGPTASFIPELFATSYRYTGSALAANLAGVAGGAL
MMAR_4238|M.marinum_M FTVALLGLYAVAASGFGPAASLIPELFATRYRYTGTALALNVAGIVGGAA
MUL_0949|M.ulcerans_Agy99 FTVALLGLYAVAASGFGPAASLIPELFATRYRYTGTALALNVAGIVGGAA
MAV_1344|M.avium_104 YVVAVLGMFGTAAVANGPTAAFVPELFATRYRYSGAAVAMNLAGIVGAAV
MAB_3416|M.abscessus_ATCC_1997 YLAAMALTYVLFGLAFGPMASFLPESYPVGTRYSGTGVAFNVAGVFGGAI
. : . . .:: : **:* .:. :. : *..
Mflv_4524|M.gilvum_PYR-GCK TPMIASSLLVDEVTSVPLVVLAMGCGAVTVLAVWRIR----ETQGVDLAG
Mvan_2899|M.vanbaalenii_PYR-1 TPLIASALHAQEVSRTPLVALAAGCGLVTVVAVWRIS----ETRGTDLTT
TH_3714|M.thermoresistible__bu APTIAAALYAATESSGSIVAYLIGVSVISAVSVCLLPGGWGRTGDQPATE
MSMEG_3586|M.smegmatis_MC2_155 SPAIATALVQRYDASTGISAYIALGGVVSALGLLLTAR--QVKAARDPAD
Mb1232|M.bovis_AF2122/97 PPVIAGALVATYG-SWAIGVMLAILALISLVCTYRLP----ETAGSALVS
Rv1200|M.tuberculosis_H37Rv PPVIAGALVATYG-SWAIGVMLAILALISLVCTYRLP----ETAGSALVS
MMAR_4238|M.marinum_M PPLIAGTLLASYG-SSAVGLMMTALVAASLLCTYLLP----ETAGAPLTA
MUL_0949|M.ulcerans_Agy99 PPLIAGTLLASYG-SSAVGLMMTALVAASLLCTYLLP----ETAGAPLTA
MAV_1344|M.avium_104 PPLLAGTLLATYG-SWAIGLMMASLVLASFVSVYLLP----ETRGAALDA
MAB_3416|M.abscessus_ATCC_1997 PPLIAGPLQNTLG-SWAVGLMLLVICAISVASISGMR----ETWAGSAR-
.* :* .* : : .
Mflv_4524|M.gilvum_PYR-GCK IDSGRNAVRTP--
Mvan_2899|M.vanbaalenii_PYR-1 V------------
TH_3714|M.thermoresistible__bu VPASVRPMVSGPD
MSMEG_3586|M.smegmatis_MC2_155 I------------
Mb1232|M.bovis_AF2122/97 R------------
Rv1200|M.tuberculosis_H37Rv R------------
MMAR_4238|M.marinum_M A------------
MUL_0949|M.ulcerans_Agy99 A------------
MAV_1344|M.avium_104 AAAAEKVAAR---
MAB_3416|M.abscessus_ATCC_1997 -------------