For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KSAARRPVLVLAALASGFIMASIDSTVVNVTTVSIGESFGSTTAMLTWIVDAYVLTYASLLLLGGSLATS FGSRRLYMLGMGVFFAASLGCAVAPSPVFLIGARAVQGIGAAMFMPSSLSLLIAQFTEPSRRARMLGLWS AMVATAAAIGPSLGGVLIGSFGWRSIFLINIPVVVVGMALTLRVVPRVSGTRQRVSPVGHLTMLGGVGAA AFLLIQGHSSGFRSPVSVIVAAVVVISGGVLLVQQRRTVNPVVPWDLFRGRVFSTVNAVGFLYSGALYGC LYAMGLFFQNARHVSPMEAGLQLLPMTMCFPLGNILYTRIHHRISNAAIMSVCLSVAGVATLLLIGVDRE TPYWLLALVLGIANSGAGLVTASMTAATVAAAGDRHANHAGAILNANRQLGVLVGVAIVGLIVEYNASWD SGLPVVAAVIASAYIAAGFAASTLIRSRESRESSSRKRADLGARHASAGAQKHVAETHCGAP*
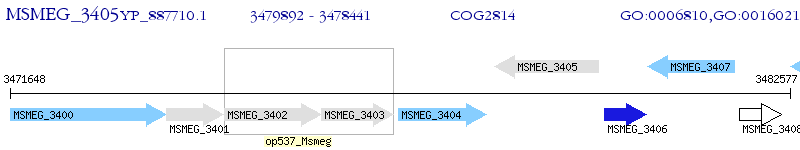
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3405 | - | - | 100% (483) | methylenomycin A resistance protein |
| M. smegmatis MC2 155 | MSMEG_5670 | - | 7e-62 | 33.73% (415) | efflux membrane protein |
| M. smegmatis MC2 155 | MSMEG_5046 | - | 2e-47 | 30.35% (402) | drug transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2361c | - | 2e-54 | 30.95% (420) | integral membrane transport protein |
| M. gilvum PYR-GCK | Mflv_1730 | - | 5e-63 | 34.62% (413) | EmrB/QacA family drug resistance transporter |
| M. tuberculosis H37Rv | Rv2333c | - | 1e-53 | 30.95% (420) | integral membrane transport protein |
| M. leprae Br4923 | MLBr_01562 | - | 2e-42 | 27.72% (404) | putative transmembrane efflux protein |
| M. abscessus ATCC 19977 | MAB_0945 | - | 6e-66 | 35.35% (413) | EmrB/QacA family drug resistance transporter |
| M. marinum M | MMAR_1146 | - | 3e-58 | 33.50% (412) | integral membrane transport protein |
| M. avium 104 | MAV_0729 | - | 1e-45 | 30.64% (408) | EmrB efflux protein |
| M. thermoresistible (build 8) | TH_3965 | - | 1e-47 | 30.24% (410) | PUTATIVE drug transporter |
| M. ulcerans Agy99 | MUL_0911 | - | 2e-58 | 33.25% (412) | integral membrane transport protein |
| M. vanbaalenii PYR-1 | Mvan_5021 | - | 7e-65 | 34.55% (411) | EmrB/QacA family drug resistance transporter |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1146|M.marinum_M --------------------------------------------MRIQPV
MUL_0911|M.ulcerans_Agy99 --------------------------------------------MRIQPV
Mb2361c|M.bovis_AF2122/97 --------------------------------------------------
Rv2333c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1730|M.gilvum_PYR-GCK -------------------------------------------------M
Mvan_5021|M.vanbaalenii_PYR-1 -------------------------------------------------M
MAB_0945|M.abscessus_ATCC_1997 ------------------------------------------MEAAVGAL
MSMEG_3405|M.smegmatis_MC2_155 -----------------------------------------------MKS
TH_3965|M.thermoresistible__bu ----------------------------VTSVDAGRSTMSEVFTAYAPRS
MLBr_01562|M.leprae_Br4923 MVTLYDHERAVHNWTAARSDRPAPSRLTQQVEPASERTSKYPTLLPSGRF
MAV_0729|M.avium_104 ----------------MLSGAMEKTRSAAFSVPLATASGPSLRDDEYPDK
MMAR_1146|M.marinum_M FRDHPIPALAVICLSVFVISVDATIVNVALPTLSRQLGADTAQLQWIVDA
MUL_0911|M.ulcerans_Agy99 FRDHPILALAVICLSVFVISVDATIVNVALPTLSRELGADTAQLQWIVDA
Mb2361c|M.bovis_AF2122/97 MNRTQLLTLIATGLGLFMIFLDALIVNVALPDIQRSFAVGEDGLQWVVAS
Rv2333c|M.tuberculosis_H37Rv MNRTQLLTLIATGLGLFMIFLDALIVNVALPDIQRSFAVGEDGLQWVVAS
Mflv_1730|M.gilvum_PYR-GCK TPRRKAIILVSCCLSLLIVSMDATIVNVAIPSIRADLSASPAQMQWVVDI
Mvan_5021|M.vanbaalenii_PYR-1 TPRRKAIILVSCCLSLLIVSMDATIVNVAIPSIRDDLSATPAQMQWVVDV
MAB_0945|M.abscessus_ATCC_1997 SERRKIVILGSCCLSLLIVSMDSTIVNVALPTIRADFGATTSQLQWVIDI
MSMEG_3405|M.smegmatis_MC2_155 AARRPVLVLAALASGFIMASIDSTVVNVTTVSIGESFGSTTAMLTWIVDA
TH_3965|M.thermoresistible__bu AAANPWHALWAMLIGFFMILVDATIVPVANLEIMADLGADYTAVIWVTSA
MLBr_01562|M.leprae_Br4923 PSGRFIAAVIVIGGMQLLATMDSTVAIVALPKIQNELSLSDAGRSWGITA
MAV_0729|M.avium_104 LDAALFRIAGVCGLACIMAVLDSTVVAVAQRTFIAQFGANQAIVSWTIAG
:: :*: :. *: : .:. *
MMAR_1146|M.marinum_M YTLVMSGLLLSAGSLSDRYGRRGWLSAGLILFALTSALAAQVNSADTLIA
MUL_0911|M.ulcerans_Agy99 YTLVMSGLLLSAGSLSDRYGRRGWLSAGLILFALTSALAAQVNSADTLVA
Mb2361c|M.bovis_AF2122/97 YSLGMAVFIMSAATLADLYGRRRWYLIGVSLFTLGSIACGLAPSIAVLTT
Rv2333c|M.tuberculosis_H37Rv YSLGMAVFIMSAATLADLDGRRRWYLIGVSLFTLGSIACGLAPSIAVLTT
Mflv_1730|M.gilvum_PYR-GCK YTLVLASLLMLSGATGDRFGRRRLFQIGLTVFALGSLACSLAPTIDTLIG
Mvan_5021|M.vanbaalenii_PYR-1 YTLVLASLLMLSGATGDRFGRRRVFQIGLAVFALGSLACSLAPTIDALIG
MAB_0945|M.abscessus_ATCC_1997 YTLGLASLLMLSGAAGDRLGRRKVFHLGLSIFAIGSLLCSVAPTVGLLIA
MSMEG_3405|M.smegmatis_MC2_155 YVLTYASLLLLGGSLATSFGSRRLYMLGMGVFFAASLGCAVAPSPVFLIG
TH_3965|M.thermoresistible__bu YLLAYAVPLLIAGRLGDRFGPRTMYLIGLAIFTAASLWCGLAGTIGMLIA
MLBr_01562|M.leprae_Br4923 YVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLCAVAWDEVTLVI
MAV_0729|M.avium_104 YMLAFATVIPITGWAADRFGTKRLFMGSVLIFTLGSLLCAVAPNILLLIL
* * . : . . * : .: :* * .. . *
MMAR_1146|M.marinum_M ARAAMGVGAAVIFPTTLGLITNIFTDPVPRAKAIGLWTAMVGVGVAVGPI
MUL_0911|M.ulcerans_Agy99 ARAAMGVGAAVIFPTTLGLITNIFTDPVPRAKAIGLWTAMVGVGVAVGPI
Mb2361c|M.bovis_AF2122/97 ARGAQGLGAAAVSVTSLALVSAAFPEAKEKARAIGIWTAIASIGTTTGPT
Rv2333c|M.tuberculosis_H37Rv ARGAQGLGAAAVSVTSLALVSAAFPEAKEKARAIGIWTAIASIGTTTGPT
Mflv_1730|M.gilvum_PYR-GCK ARFLQGVGGSMLNPVALSIISQIFTKPVERARALGIWGGVTGISMAAGPI
Mvan_5021|M.vanbaalenii_PYR-1 ARLLQGIGGSMLNPVALSIISQIFTQPVERARALGIWGGVTGISMAAGPI
MAB_0945|M.abscessus_ATCC_1997 ARLLQAVGGSMLNPVALSIISQVFTGRVERARALGFWGAVVGISMALGPI
MSMEG_3405|M.smegmatis_MC2_155 ARAVQGIGAAMFMPSSLSLLIAQFTEPSRRARMLGLWSAMVATAAAIGPS
TH_3965|M.thermoresistible__bu ARVLQGLGAALLTPQTLSMITRIFPAD-RRGVPMGVWGATAGVATLIGPL
MLBr_01562|M.leprae_Br4923 ARLLQGIGSAIASPTGLALVATTFSKGSARNTATAVFAAMTAIGSVMGLV
MAV_0729|M.avium_104 FRVVQGVGGGMLLPLSFVILTR-EAGPKRVGRLMAVGGIPILLGPIGGPI
* .:*.. : :: . .. . *
MMAR_1146|M.marinum_M TGGWLLEHFSWGSIFLVNVPIAVAAMAGAILFVPTSRDPAAPRVDVPGLM
MUL_0911|M.ulcerans_Agy99 TGGWLLEHFSWGSIFLVNVPIAVAAMAGAILFVPTSRDPAAPRVDVPGLM
Mb2361c|M.bovis_AF2122/97 LGGLLVDQWGWRSIFYVNLPMGALVLFLTLCYVEESCNERARRFDLSGQL
Rv2333c|M.tuberculosis_H37Rv LGGLLVDQWGWRSIFYVNLPMGALVLFLTLCYVEESCNERARRFDLSGQL
Mflv_1730|M.gilvum_PYR-GCK VGGLLIETVGWRSVFWINLPICAAAIILTAVFVPESKSQTMRNVDPIGQG
Mvan_5021|M.vanbaalenii_PYR-1 VGGLLIDTIGWRSVFWINLPICAAAILLTAIFVPESKSQTMRNVDPIGQG
MAB_0945|M.abscessus_ATCC_1997 VGGLLIQWVGWRAVFWINLPVCVLAFVLTAVFVPETKSATMRDIDPVGQA
MSMEG_3405|M.smegmatis_MC2_155 LGGVLIGSFGWRSIFLINIPVVVVGMALTLRVVPRVSG-TRQRVSPVGHL
TH_3965|M.thermoresistible__bu AGGVLVDGLGWQWIFIVNVPIGIAGLVLGARLMP-RLPRQDQGFDVLGMV
MLBr_01562|M.leprae_Br4923 VGGALTEVS-WRLAFLVNVPIGLVMIYLARTALHETHKERMK-LDAAGAI
MAV_0729|M.avium_104 LGGWLIGAYGWKWIFLINLPIGLTAFALAALLFPKDRSAPSEALDITGAL
** * * * :*:*: : . .. *
MMAR_1146|M.marinum_M LSAVGITALVYTIIEAPNWGWTSTRASGGFIAAAIVLVGFALWER--RSS
MUL_0911|M.ulcerans_Agy99 LSAVGITALVYTIIEAPNWGWTSTRASGGFIAAAIVLVGFALWER--RSS
Mb2361c|M.bovis_AF2122/97 LFIVAVGALVYAVIEGPQIGWTSVQTIVMLWTAAVGCALFVWLER--RSS
Rv2333c|M.tuberculosis_H37Rv LFIVAVGALVYAVIEGPQIGWTSVQTIVMLWTAAVGCALFVWLER--RSS
Mflv_1730|M.gilvum_PYR-GCK LGILFLFGTVFTLIEGPVLGWTNPRVLAIAAVAVVALIAFLRFES--RRH
Mvan_5021|M.vanbaalenii_PYR-1 LAIMFLFGTVFTLIEGPVLGWTNPRVVVAAVVVALALVAFLRFES--RRH
MAB_0945|M.abscessus_ATCC_1997 LAVLFLFGIVFALIEGPSLGWATPGLIVVLAVALAAFAVFLRYES--RRR
MSMEG_3405|M.smegmatis_MC2_155 TMLGGVGAAAFLLIQGHSSGFRSPVSVIVAAVVVISGGVLLVQQR--RTV
TH_3965|M.thermoresistible__bu LSAVGLFLIVFALQEAQTHRWAPWIWGTLAGGAGVMAMFVIWQSA--NRR
MLBr_01562|M.leprae_Br4923 LATLACTAAVFAFSMGPEKGWISLTTIGSGLVALVAAIAFAVVER--TAE
MAV_0729|M.avium_104 LLSPGVAIFLCGVCSIPGRHTVADRYVLVPALVGLVLIAAFILHAWYRTE
. .
MMAR_1146|M.marinum_M HPMLDVSVFANRRFSGGSLAVTAGFLTLFGFIFVITQYFQFIKDYTALQT
MUL_0911|M.ulcerans_Agy99 HPMLDVSVFANRRFSGGSLAVTAGFLTLFGFIFVITQYFQFIKDYTALQT
Mb2361c|M.bovis_AF2122/97 NPMMDLTLFRDTSYALAIATICTVFFAVYGMLLLTTQFLQNVRGYTPSVT
Rv2333c|M.tuberculosis_H37Rv NPMMDLTLFRDTSYALAIATICTVFFAVYGMLLLTTQFLQNVRGYTPSVT
Mflv_1730|M.gilvum_PYR-GCK DPFLDLRFFRSIPFTTATINAISAFAAWGAFLFMMSLYLQGERGFSAMQT
Mvan_5021|M.vanbaalenii_PYR-1 DPFLDLRFFRSIPFTTATVIAISAFAAWGAFLFLMSLYLQGERGFSAMHT
MAB_0945|M.abscessus_ATCC_1997 HPFIDLRFFRSIPFASATLTAICACMAWSAFLFTMSLYLQGQRHFSAMHT
MSMEG_3405|M.smegmatis_MC2_155 NPVVPWDLFRGRVFSTVNAVGFLYSGALYGCLYAMGLFFQNARHVSPMEA
TH_3965|M.thermoresistible__bu HPLVPLQIFADRNFMLANVGVATIGFVVTAMVLPAMFFAQAVCGLSPTGS
MLBr_01562|M.leprae_Br4923 NPVVPFDLFRDRNRLVTFIAIFLAGGVIFSLTVSIGLYIQDILGYSALRA
MAV_0729|M.avium_104 HPLIDLRLFRNPVVTQVNVTLLVFAAASVGVGLLVPSYFQIVGHETPMQS
.*.: .* . . . : * :. :
MMAR_1146|M.marinum_M GVRLLPVALSIALAAILGPRLVERVGTTAVVAAGLAVFAA-ALAWASTAD
MUL_0911|M.ulcerans_Agy99 GVRLLPVAFSIALAAILGPRLVERVGTTAVVAAGLAVFAA-ALAWASTAD
Mb2361c|M.bovis_AF2122/97 GLMILPFSAAVAIVSPLVGHLVGRIGAR-VPILAGLCMLM-LGLLMLIFS
Rv2333c|M.tuberculosis_H37Rv GLMILPFSAAVAIVSPLVGHLVGRIGAR-VPILAGLCMLM-LGLLMLIFS
Mflv_1730|M.gilvum_PYR-GCK GLIYLPIALGALFCSPLSGRLVGRYGARPSLVAAGILITT-AATLLTLLT
Mvan_5021|M.vanbaalenii_PYR-1 GLIYLPIAIGALLFSPLSGRLVGRYGARPSLVIAGVTITA-AATMLTFLT
MAB_0945|M.abscessus_ATCC_1997 GLIYLPIAVGALVFSPISGRMVGRFGARPSLLAAGTLFLT-ASLLLTRVS
MSMEG_3405|M.smegmatis_MC2_155 GLQLLPMTMCFPLGNILYTRIHHRISNAAIMSVCLSVAGV-ATLLLIGVD
TH_3965|M.thermoresistible__bu ALLIAPMAVASGVLAPMAGRIVDRSHPTPVVGFGFSVVAVGLTWLAVEMA
MLBr_01562|M.leprae_Br4923 GIGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGWLLMVAMLYGWAFMN
MAV_0729|M.avium_104 GLHMLPIGVGAVLTMPLGGAVMDKHGPGKIVLTGLPLMAVGLAVFTYGVA
.: *. . : : :
MMAR_1146|M.marinum_M ATTPYTQIALQMLLLG-GGLGLTTAPATEAIMGSLPADRAGVGSAVNDTT
MUL_0911|M.ulcerans_Agy99 ATTPYTQITLQMLLLG-GGLGLTTAPATEAIMGSLPADRAGVGSAVNDTT
Mb2361c|M.bovis_AF2122/97 EHRSSALVLVGLGLCG-SGVALCLTPITTVAMTAVPAERAGMASGIMSAQ
Rv2333c|M.tuberculosis_H37Rv EHRSSALVLVGLGLCG-SGVALCLTPITTVAMTAVPAERAGMASGIMSAQ
Mflv_1730|M.gilvum_PYR-GCK ATTPVWHLLIVFAVFG-VGFSMVNAPITNAAVSGMPLDRAGAASAVTSTS
Mvan_5021|M.vanbaalenii_PYR-1 ATTPVWSLLMVFAVFG-IGFSMVNAPVTNAAVSGMPLDRAGAASAVTSTS
MAB_0945|M.abscessus_ATCC_1997 EATPIWELLVTFAIFG-AGFAMVNAPITNAAVSGMPLDRAGAASAVTSTS
MSMEG_3405|M.smegmatis_MC2_155 RETPYWLLALVLGIAN-SGAGLVTASMTAATVAAAGDRHANHAGAILNAN
TH_3965|M.thermoresistible__bu PTTPIWRLVLPFVAVG-VGMAFIWSPLATTATRQLPLHLSGAGSGVYNTT
MLBr_01562|M.leprae_Br4923 RGVPYFPNLLAPIVVGGIGIGMAVVPLTLSAIAGVGFDRIGPVSAIALML
MAV_0729|M.avium_104 RQAAYSPVLVCGLAIMGLGIGLTTTPLSAALMQALAPHQVARGTTLISVN
. : * .: . : :
MMAR_1146|M.marinum_M RELGGTLGVAIAGSVFASVYSGQL---------------------GSAST
MUL_0911|M.ulcerans_Agy99 RELGGTLGVAIAGSVFASVYSGQL---------------------GSASA
Mb2361c|M.bovis_AF2122/97 RAIGSTIGFAVLGSVLAAWLSATL---------------------EPHLE
Rv2333c|M.tuberculosis_H37Rv RAIGSTIGFAVLGSVLAAWLSATL---------------------EPHLE
Mflv_1730|M.gilvum_PYR-GCK RQVGVSIGVALCGSVAGAAMAGSG---------------------TDFAT
Mvan_5021|M.vanbaalenii_PYR-1 RQVGVSIGVALCGSLAGSAMATGG---------------------ADFAA
MAB_0945|M.abscessus_ATCC_1997 RQVGVSIGVALCGSIG----------------------------------
MSMEG_3405|M.smegmatis_MC2_155 RQLGVLVGVAIVGLIVEYN-----------------------------AS
TH_3965|M.thermoresistible__bu RQIGSVLGSAGMAAFLTGRLSAELPFGATAVPAGEQTRVLPEFLHAPYAA
MLBr_01562|M.leprae_Br4923 QSLGGPLVLAVIQAVITSRTLYLG-------------------------G
MAV_0729|M.avium_104 QQVGGSIGAALMAVILTNQFN-----------------------------
: :* : * .
MMAR_1146|M.marinum_M LATLPSDVRSAMKGSMAVAHKVIDQLPAGPAAGVRDAVNHAFLDGLQVAT
MUL_0911|M.ulcerans_Agy99 LATLPSDVRSAMKGSMAVAHKVIDQLPAGPAAGVRDAVNHAFLDGLQVAT
Mb2361c|M.bovis_AF2122/97 RAVPDPVQRHVLA--EIIIDSANPRAHVGGIVPRRHIEHRDPVAIAEEDF
Rv2333c|M.tuberculosis_H37Rv RAVPDPVQRHVLA--EIIIDSANPRAHVGGIVPRRHIEHRDPVAIAEEDF
Mflv_1730|M.gilvum_PYR-GCK AARPLWFVCIALG--LVILAVG---FFSTSQRAVRSAERLAPLVAGAPDP
Mvan_5021|M.vanbaalenii_PYR-1 AARPLWFVCMALG--LVILGLG---LFATSQRALRSAERLAPLVSGAPEP
MAB_0945|M.abscessus_ATCC_1997 -TPALWFMCAGLG--LLIATLG---IVSTSPAAVRSAQRLAPLIEGT-TA
MSMEG_3405|M.smegmatis_MC2_155 WDSGLPVVAAVIASAYIAAGFAASTLIRSRESRESSSRKRADLGARHASA
TH_3965|M.thermoresistible__bu AMAQSMLLPAFVALFGVLAAIFLAGIVLSTPQPDADAQYLDEHDDYLVIE
MLBr_01562|M.leprae_Br4923 TTGPVKFMNDAQLRALDHGYTYGLLWVAGAVVIVGVAALFIGYTPEQVAY
MAV_0729|M.avium_104 -RNPALMAANEAAGMHPVTGKRGLPVDPSTVPRPAMTPELAGHVSHHLSH
MMAR_1146|M.marinum_M LVCAGIALAAAIVVGWLLPARAGQPTPAQTEPALAQA-------------
MUL_0911|M.ulcerans_Agy99 LVCAGIALAAAIVVGWLLPARAGQPTPAQTEPALAQV-------------
Mb2361c|M.bovis_AF2122/97 IEGIRVALLVATATLAVVFLAGWRWFPRDVQTAGSDAKR--EAAYSDDRR
Rv2333c|M.tuberculosis_H37Rv IEGIRVALLVATATLAVVFLAGWRWFPRDVHTAGSDLSERLPTAMTVECA
Mflv_1730|M.gilvum_PYR-GCK VNGAHVR-------------------------------------------
Mvan_5021|M.vanbaalenii_PYR-1 VGGAHVR-------------------------------------------
MAB_0945|M.abscessus_ATCC_1997 REMSRVG-------------------------------------------
MSMEG_3405|M.smegmatis_MC2_155 GAQKHVAETHCGAP------------------------------------
TH_3965|M.thermoresistible__bu LARPDPNPRTDIAVTAVIPALRPTLRPTLRPSLGPPGGPVVQPNNTAALR
MLBr_01562|M.leprae_Br4923 AQEVKEAVDAGEL-------------------------------------
MAV_0729|M.avium_104 AYTAVFVLAVVLVACTIIPASFLPRKPPSPPAGD----------------
MMAR_1146|M.marinum_M ---------------
MUL_0911|M.ulcerans_Agy99 ---------------
Mb2361c|M.bovis_AF2122/97 VRG------------
Rv2333c|M.tuberculosis_H37Rv VSHMPGATWCRLWPA
Mflv_1730|M.gilvum_PYR-GCK ---------------
Mvan_5021|M.vanbaalenii_PYR-1 ---------------
MAB_0945|M.abscessus_ATCC_1997 ---------------
MSMEG_3405|M.smegmatis_MC2_155 ---------------
TH_3965|M.thermoresistible__bu ---------------
MLBr_01562|M.leprae_Br4923 ---------------
MAV_0729|M.avium_104 ---------------