For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTGLDGLNPTPGALVLGLVQARVPVIKEPGDLKSTAERLAGMVVGAKKGMPSIDLVVMPEYSINGLDPDT WLDDSLLCDRNGPEMTLLADACRDAGVWGCFSIMERNRGGAPWNSGIIIDDHGEERLYYRKMHPWVPAEP WAPGDLGVPVCDGPSGSRLALIICHDGMLPEMAREAAYKGANVILRTAGYTYPIQHSWRITNQTNAFHNL AYTASVALAGPDQNNIWSQGEAMVCDVDGTILVSGDGTPDRIVTAEVVPSRADEARRTWGVENNIYQLGH RGYTAVTGGAQDCPYTFMQDLVAGTYRLPWEDEVRHVDGTGAGWGAPETVRAK
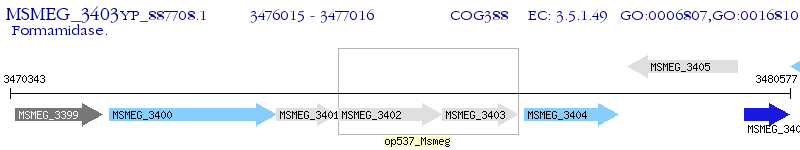
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3403 | amiF | - | 100% (333) | formamidase |
| M. smegmatis MC2 155 | MSMEG_6733 | - | 1e-11 | 29.32% (266) | hydrolase, carbon-nitrogen family protein |
| M. smegmatis MC2 155 | MSMEG_0443 | - | 9e-07 | 25.30% (249) | hydrolase, carbon-nitrogen family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0095 | - | 3e-05 | 25.58% (215) | Nitrilase/cyanide hydratase and apolipoprotein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0819 | - | 3e-13 | 27.67% (253) | carbon-nitrogen hydrolase family protein |
| M. marinum M | MMAR_4512 | - | 3e-05 | 25.91% (220) | amidohydrolase |
| M. avium 104 | MAV_4670 | - | 5e-06 | 24.86% (173) | carbon-nitrogen hydrolase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3160 | - | 1e-09 | 26.22% (267) | Nitrilase/cyanide hydratase and apolipoprotein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0095|M.gilvum_PYR-GCK ---------MRSGMRIALAQIQS-STDPAANLTLVEEYTRRAVDAGAS--
MAV_4670|M.avium_104 -------------MRIALAQILS-GTDPAANLALVGEYTRRAAGAGAR--
MAB_0819|M.abscessus_ATCC_1997 ---------MTGRLRLAVVQCSSQAADVQANLARLAETAHAAAAGDAQ--
MMAR_4512|M.marinum_M ---------MHQVETELVVAVAQPRTEMGAVAANVDAHAALIRRANAR--
MSMEG_3403|M.smegmatis_MC2_155 MTGLDGLNPTPGALVLGLVQARVPVIKEPGDLKSTAERLAGMVVGAKKGM
Mvan_3160|M.vanbaalenii_PYR-1 ----------MAPSTYTLAMAQPRRIAGPDGSANVAHAVRLVEQAAGAD-
:. .
Mflv_0095|M.gilvum_PYR-GCK ----LVLFPEATMVRFGV---PLGGVAEPFDGPWANAVRDIAARAGIVVV
MAV_4670|M.avium_104 ----LVVFPEATMCRFGV---PLAPIAEPVDGPWADGVRRIATEANVTVI
MAB_0819|M.abscessus_ATCC_1997 ----LVVFPEMYLTGYNIGEWDIRDLAQSPDGPAMQFIRQTACDADMHIC
MMAR_4512|M.marinum_M ----LVVFPELSLTGYQL-----KVAPVDVAGPVVDRLVLACAATGCSVL
MSMEG_3403|M.smegmatis_MC2_155 PSIDLVVMPEYSINGLDPDTWLDDSLLCDRNGPEMTLLADACRDAGVWGC
Mvan_3160|M.vanbaalenii_PYR-1 ----LVCFPEFSPG---------PVRAHDTFYDAGPALAAAARACDVNVV
** :** : . .
Mflv_0095|M.gilvum_PYR-GCK AGMFVPSDDGRSFNTLLATGPG--VEARYDKIHLYDAFGFLESKTVAPGR
MAV_4670|M.avium_104 AGMFTPSGDGRVKNTLLVANSDDQAVTHYDKIHLYDAFGFTESRTVAPGR
MAB_0819|M.abscessus_ATCC_1997 VGYPERASDGQIYNSALICDPRGRVVLNYRKSHLFGDT--ERNVFTRPGP
MMAR_4512|M.marinum_M VGAPVEDRGRRYIAMVLVDSSG--AEVVYRKTHL----GAAEKARFRPGP
MSMEG_3403|M.smegmatis_MC2_155 FSIMERNRGGAPWNSGIIIDDHGEERLYYRKMHP-----WVPAEPWAPG-
Mvan_3160|M.vanbaalenii_PYR-1 WSRTERCDDGRNRLVVYVTGRDGDPVLRYARTHPATIPPGEDGELVSPAD
. . . * : * *.
Mflv_0095|M.gilvum_PYR-GCK EPVVISVDTESGPVTVGLTLCYDIRFPELYVELADRGAEIITAHASWGTG
MAV_4670|M.avium_104 EPVVVGVDG----VRVGLSVCYDIRFPELYTELARRGAQLIAVCASWGAG
MAB_0819|M.abscessus_ATCC_1997 QLPLAVVHG----VSVGLLICYDVEFPENVRALAMARADVVVVPTAN--M
MMAR_4512|M.marinum_M GPRTIDLDG----WRIGLGICRDTGVPEHVRGTARLGVDLYACGVVHHDW
MSMEG_3403|M.smegmatis_MC2_155 DLGVPVCDGPSG-SRLALIICHDGMLPEMAREAAYKGANVILRTAGY--T
Mvan_3160|M.vanbaalenii_PYR-1 TFGNFEVDG----IPMGIVVCSEMWTPEIARIVALRGAEIILSPAGGG-F
. :.: :* : ** * .:: .
Mflv_0095|M.gilvum_PYR-GCK PGKLEQWTLLARARAIDTSSVVAAVGQAYPG--DEIAALG-PTGVGGSLV
MAV_4670|M.avium_104 PGKLDQWTLLARARALDSMSYIAAAGQADPG--GSLSASGAPTGVGGSLV
MAB_0819|M.abscessus_ATCC_1997 EPFHAVCETVVPARAYENQLYVAYANYCGNH--DGLAYCG------GSSI
MMAR_4512|M.marinum_M ELGEQRHRARTIAATCDAPVAMASFAGPTGG--GYLHTAG------HSAI
MSMEG_3403|M.smegmatis_MC2_155 YPIQHSWRITNQTNAFHNLAYTASVALAGPDQ-NNIWSQG------EAMV
Mvan_3160|M.vanbaalenii_PYR-1 TSLTRNWQIIVSARAIENLCYIGLTNNLWGDEQGAAMITG-----PEHPL
: : . . . * :
Mflv_0095|M.gilvum_PYR-GCK ASALGEVLASAGDDP-ELLVCDVDLEAARKARQTVAVMQNRSGFAHRGKA
MAV_4670|M.avium_104 ASPLGEVVASAGAQP-QLVLADIDVDRVAQARKTIAALSNRSDFAQVGRA
MAB_0819|M.abscessus_ATCC_1997 VGPDGTALARAGEGV-GLLVADVDRAVIDRTRSRHCFLEDR-----VARA
MMAR_4512|M.marinum_M WSATSTLVAEAGAGPDQIAHTTIGRPPPGRAGDDHTPKTA----------
MSMEG_3403|M.smegmatis_MC2_155 CDVDGTILVSGDGTPDRIVTAEVVPSRADEARRTWGVENNIYQLGHRGYT
Mvan_3160|M.vanbaalenii_PYR-1 AHAGRSELVTATVDLDRLRWLRAHDDSMAEPKAFDSIPGLLRARRPELYG
:. . : ..
Mflv_0095|M.gilvum_PYR-GCK ESRG---------------------------------------------
MAV_4670|M.avium_104 ESVG---------------------------------------------
MAB_0819|M.abscessus_ATCC_1997 ISP----------------------------------------------
MMAR_4512|M.marinum_M -------------------------------------------------
MSMEG_3403|M.smegmatis_MC2_155 AVTGGAQDCPYTFMQDLVAGTYRLPWEDEVRHVDGTGAGWGAPETVRAK
Mvan_3160|M.vanbaalenii_PYR-1 ELVAPQADAYDYANGQA--------------------------------