For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LVSHSKDSAETPTLAESVYQRIRRNILEGRIMPGTRVSIRSLAESVGVSTMPTREALKRLSFEGLVEYDR RAVTISTLSPKAIRELFTIRLRLELLATEWALPRLDAETTVALRSVLDRMIVPGISAITWRELNREFHQT FYSCGDSRYLLELIHNIWDRIQPYMAIYASAMHDFTEADRQHEHMYQLMLARDLDGLLDATTRHLEHTAE AIVSALGADTTTGRGNNVNYEELSISSFHRLVESGGATSADLTAWYLDRIATLDSIDNPDGPQLNSVVTV NPAALDEARALDEKYARTGTLVGPLHGVPILIKDQGETKGIPTAFGSTAFADYIPDTDATVVDRLRKAGA VILGKTAMCDFAAGWFSFSSRTDHTKNPYDLDRETGGSSAGTAAAVTANLCLVGIGEDTGGSIRLPSSFT NLFGLRVTTGLVSRTGFSPLVHFQDTPGPMARNVSDLAAVLDVIVGYDPTDSYTALATSGPEVGDYGEAL DGVDADTLSGFRVGVLTDAFGAGDDQELTNSVVRAAIDRLRHHGTGVVDGIVLGNLETWVAETSLYTIQS KFDLEKFLGSRRHTGPTTIREIVDHGDFHPLTDLLADIADGPSNPEDHPEYFKKRTRQEDWRRTLLAKMA DAEIDFLVYPTVQVPAPTRADLAAKRWTALNFPTNTVIASQTSLPAMSIPVGFTDSGLPVGLEVVGRPLT ERALLRFARAWELAEAPRRQPQIEASKARVTM
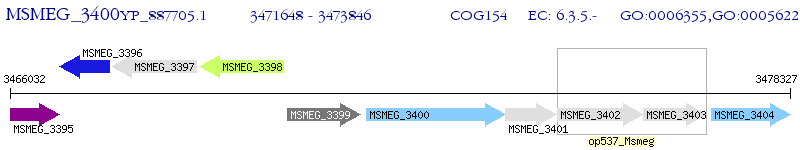
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3400 | - | - | 100% (732) | glutamyl-tRNA(Gln) amidotransferase subunit A |
| M. smegmatis MC2 155 | MSMEG_0485 | - | 2e-47 | 33.41% (434) | amidase |
| M. smegmatis MC2 155 | MSMEG_1090 | - | 2e-47 | 31.20% (468) | amidase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3409 | amiD | 3e-43 | 32.40% (500) | amidase AmiD |
| M. gilvum PYR-GCK | Mflv_4246 | gatA | 8e-43 | 31.30% (492) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. tuberculosis H37Rv | Rv3375 | amiD | 3e-43 | 32.40% (500) | amidase AmiD |
| M. leprae Br4923 | MLBr_01702 | gatA | 7e-43 | 30.54% (501) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. abscessus ATCC 19977 | MAB_3341c | gatA | 2e-44 | 31.01% (487) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. marinum M | MMAR_0937 | gatA_1 | 1e-54 | 33.14% (516) | peptide amidase, GatA_1 |
| M. avium 104 | MAV_3859 | gatA | 4e-40 | 30.46% (499) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. thermoresistible (build 8) | TH_2639 | - | 2e-41 | 31.20% (484) | PUTATIVE glutamyl-tRNA (Gln) amidotransferase (subunit A), GatA |
| M. ulcerans Agy99 | MUL_0689 | gatA_1 | 5e-52 | 32.63% (524) | peptide amidase, GatA_1 |
| M. vanbaalenii PYR-1 | Mvan_2115 | gatA | 3e-39 | 30.45% (486) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_01702|M.leprae_Br4923 --------------------------------------------------
MAV_3859|M.avium_104 --------------------------------------------------
TH_2639|M.thermoresistible__bu --------------------------------------------------
Mflv_4246|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2115|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3341c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3409|M.bovis_AF2122/97 --------------------------------------------------
Rv3375|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0937|M.marinum_M --------------------------------------------------
MUL_0689|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_3400|M.smegmatis_MC2_155 MVSHSKDSAETPTLAESVYQRIRRNILEGRIMPGTRVSIRSLAESVGVST
MLBr_01702|M.leprae_Br4923 --------------------------------------------------
MAV_3859|M.avium_104 --------------------------------------------------
TH_2639|M.thermoresistible__bu --------------------------------------------------
Mflv_4246|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2115|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3341c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3409|M.bovis_AF2122/97 --------------------------------------------------
Rv3375|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0937|M.marinum_M --------------------------------------------------
MUL_0689|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_3400|M.smegmatis_MC2_155 MPTREALKRLSFEGLVEYDRRAVTISTLSPKAIRELFTIRLRLELLATEW
MLBr_01702|M.leprae_Br4923 --------------------------------------------------
MAV_3859|M.avium_104 --------------------------------------------------
TH_2639|M.thermoresistible__bu --------------------------------------------------
Mflv_4246|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2115|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3341c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3409|M.bovis_AF2122/97 --------------------------------------------------
Rv3375|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0937|M.marinum_M --------------------------------------------------
MUL_0689|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_3400|M.smegmatis_MC2_155 ALPRLDAETTVALRSVLDRMIVPGISAITWRELNREFHQTFYSCGDSRYL
MLBr_01702|M.leprae_Br4923 --------------------------------------------------
MAV_3859|M.avium_104 --------------------------------------------------
TH_2639|M.thermoresistible__bu --------------------------------------------------
Mflv_4246|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2115|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3341c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3409|M.bovis_AF2122/97 --------------------------------------------------
Rv3375|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0937|M.marinum_M --------------------------------------------------
MUL_0689|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_3400|M.smegmatis_MC2_155 LELIHNIWDRIQPYMAIYASAMHDFTEADRQHEHMYQLMLARDLDGLLDA
MLBr_01702|M.leprae_Br4923 ----------------------MIGSLHEIIRLDASTLAAKIVAKELSSV
MAV_3859|M.avium_104 --------------------------MNEIIRSDAATLAARIAAKELSSV
TH_2639|M.thermoresistible__bu --------------------MSQGKSTDDLIRLDAATLGAKIAAKEISST
Mflv_4246|M.gilvum_PYR-GCK --------------------------MSELIKLDAATLADKISSKEVSSA
Mvan_2115|M.vanbaalenii_PYR-1 --------------------------MTDLIKLDAATLAAKIAAREVSAT
MAB_3341c|M.abscessus_ATCC_199 --------------------------MTDLIRQDAAALAALIHSGEVSSV
Mb3409|M.bovis_AF2122/97 -------------MTDADSAVPPRLDEDAISKLELTEVADLIRTRQLTSA
Rv3375|M.tuberculosis_H37Rv -------------MTDADSAVPPRLDEDAISKLELTEVADLIRTRQLTSA
MMAR_0937|M.marinum_M ---------------------------MELPEFTIAETQTAFERGEWTAA
MUL_0689|M.ulcerans_Agy99 ---------------------------MELPEFTIAETQTAFERGEWTAA
MSMEG_3400|M.smegmatis_MC2_155 TTRHLEHTAEAIVSALGADTTTGRGNNVNYEELSISSFHRLVESGGATSA
. : . ::.
MLBr_01702|M.leprae_Br4923 EITQACLDQIEATD------DTYRAFLHVAAEKALSAAAAVDK----AVA
MAV_3859|M.avium_104 EATQACLDQIAATD------ERYHAFLHVAADEALGAAAVVDE----AVA
TH_2639|M.thermoresistible__bu ELTQACLDRIAATD------DRYNAFLHVAANKALSAAARVDA----AVA
Mflv_4246|M.gilvum_PYR-GCK EVTQAFLDQIAATD------GDYHAFLHVGAEQALAAAQTVDR----AVA
Mvan_2115|M.vanbaalenii_PYR-1 EVTQACLDQIAATD------AEYHAFLHVAGDQALAAAATVDKAIHEATA
MAB_3341c|M.abscessus_ATCC_199 EVTRAHLDQIASTD------ETYRAFLHVAGEQALAAAKDVDD----AIA
Mb3409|M.bovis_AF2122/97 EVTESTLRRIERLD------PQLKSYAFVMPETALAAARAADAD-----I
Rv3375|M.tuberculosis_H37Rv EVTESTLRRIERLD------PQLKSYAFVMPETALAAARAADAD-----I
MMAR_0937|M.marinum_M GLTDCYLRRIREID----QSGPMLRSIIEVNPDALAIAEALDA----ERS
MUL_0689|M.ulcerans_Agy99 GLTDCYLRRIREID----QSGPMLRSIIEVNSDALAIAEALDA----ERS
MSMEG_3400|M.smegmatis_MC2_155 DLTAWYLDRIATLDSIDNPDGPQLNSVVTVNPAALDEARALDE----KYA
* * :* * ** * *
MLBr_01702|M.leprae_Br4923 AGGQLSSTLAGVPLALKDVFTT-VDMPTTCGSKILQGWHSPYDATVTTRL
MAV_3859|M.avium_104 AGERLPSPLAGVPLALKDVFTT-VDMPTTCGSKILQGWRSPYDATVTTKL
TH_2639|M.thermoresistible__bu AGEELPSPLAGVPLALKDVFTT-IDMPTTCGSKILDGWQAPYDATVTVAL
Mflv_4246|M.gilvum_PYR-GCK AGEHLPSPLAGVPLALKDVFTT-TDMPTTCGSKVLEGWTSPYDATVTAKL
Mvan_2115|M.vanbaalenii_PYR-1 AGERLPSPLAGVPLALKDVFTT-TDMPTTCGSKILEGWTSPYDATVTAKL
MAB_3341c|M.abscessus_ATCC_199 AGS-VPSGLAGVPLALKDVFTT-RDMPTTCGSKILENWVPPYDATVTARL
Mb3409|M.bovis_AF2122/97 ARGHYEGVLHGVPIGVKDLCYT-VDAPTAAGTTIFRDFRPAYDATVVARL
Rv3375|M.tuberculosis_H37Rv ARGHYEGVLHGVPIGVKDLCYT-VDAPTAAGTTIFRDFRPAYDATVVARL
MMAR_0937|M.marinum_M -GGRIRGALHGVPVVIKDSIDTGDKMATTAGSLALEGNIATRDAFVVKQL
MUL_0689|M.ulcerans_Agy99 -GGRIRGALHGVPVVIKDSIDTGDKMATTAGSLALEGNIATRDAFVVKQL
MSMEG_3400|M.smegmatis_MC2_155 RTGTLVGPLHGVPILIKDQGET-KGIPTAFGSTAFADYIPDTDATVVDRL
. * ***: :** * .*: *: : . . ** *. *
MLBr_01702|M.leprae_Br4923 RAAGIPILGKTNMDEFAMGSS-----------TENSAYGPTRNPWNVDRV
MAV_3859|M.avium_104 RAAGIPILGKTNMDEFAMGSS-----------TENSAYGPTRNPWNVERV
TH_2639|M.thermoresistible__bu RAAGIPILGKTNLDEFAMGSS-----------TENSAFGPTRNPWDVNRV
Mflv_4246|M.gilvum_PYR-GCK RSAGIPILGKTNMDEFAMGSS-----------TENSAYGPTRNPWDTERV
Mvan_2115|M.vanbaalenii_PYR-1 RAAGIPILGKTNMDEFAMGSS-----------TENSAYGPTRNPWNVDCV
MAB_3341c|M.abscessus_ATCC_199 RGAGIPILGKTNMDEFAMGSS-----------TENSAYGPTRNPWDVERV
Mb3409|M.bovis_AF2122/97 RAAGAVIIGKLAMTEGAYLG-------------YHPSLPTPVNPWDPTAW
Rv3375|M.tuberculosis_H37Rv RAAGAVIIGKLAMTEGAYLG-------------YHPSLPTPVNPWDPTAW
MMAR_0937|M.marinum_M RDAGAVILGKANMSEWG------YMRSTRPCSGWSSRGGQVRNPYVLDRS
MUL_0689|M.ulcerans_Agy99 RDAGAVILGKANMSEWGNMSEWGYMRSTRPCSGWSSRGGQVRNPYVLDRS
MSMEG_3400|M.smegmatis_MC2_155 RKAGAVILGKTAMCDFAAG-----------WFSFSSRTDHTKNPYDLDRE
* ** *:** : : . . **:
MLBr_01702|M.leprae_Br4923 PGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTATVGVKPTYGTVSR
MAV_3859|M.avium_104 PGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTATVGVKPTYGTVSR
TH_2639|M.thermoresistible__bu PGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTATVGVKPTYGTVSR
Mflv_4246|M.gilvum_PYR-GCK PGGSGGGSAAALAAFQAPLAIGTDTGGSIRQPAALTATVGVKPTYGTVSR
Mvan_2115|M.vanbaalenii_PYR-1 PGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTATVGVKPTYGTVSR
MAB_3341c|M.abscessus_ATCC_199 PGGSGGGSAAALAAFQAPLAIGTDTGGSIRQPAALTATVGVKPTYGTVSR
Mb3409|M.bovis_AF2122/97 AGVSSSGCGVATAAGLCFGSIGSDTGGSIRFPTSMCGVTGIKPTWGRVSR
Rv3375|M.tuberculosis_H37Rv AGVSSSGCGVATAAGLCFGSIGSDTGGSIRFPTSMCGVTGIKPTWGRVSR
MMAR_0937|M.marinum_M PLGSSSGSAVAVAANLCVAALGAEVDGSIVRPASSNSIVGLKPTVGLLSR
MUL_0689|M.ulcerans_Agy99 PLGSSSSSAVAVAANLCVAALGAEVDGSIVRPASSNSIVGLKPTVGLLSR
MSMEG_3400|M.smegmatis_MC2_155 TGGSSAGTAAAVTANLCLVGIGEDTGGSIRLPSSFTNLFGLRVTTGLVSR
. *... ..* :* . .:* :..*** *:: *:: * * :**
MLBr_01702|M.leprae_Br4923 YGLVACASSLDQGGPCARTVLDTALLHAVIAGHDARD----STSVEAAVP
MAV_3859|M.avium_104 YGLVACASSLDQGGPCARTVLDTALLHQVIAGHDIRD----STSVDAPVP
TH_2639|M.thermoresistible__bu YGLVACASSLDQGGPCGRTVLDTALLHQVIAGHDAKD----STSLTAPVP
Mflv_4246|M.gilvum_PYR-GCK YGLVACASSLDQGGPCARTVLDTALLHQVIAGHDPLD----STSVEAPVP
Mvan_2115|M.vanbaalenii_PYR-1 YGLIACASSLDQGGPCARTVLDTALLHQVIAGHDPRD----STSVNAAVP
MAB_3341c|M.abscessus_ATCC_199 FGLVACASSLDQGGPCARTVLDTALLHQVIAGHDPRD----STSVDEPVP
Mb3409|M.bovis_AF2122/97 HGVVELAASYDHVGPITRSAHDAAVLLSVIAGSDIHD----PSCSAEPVP
Rv3375|M.tuberculosis_H37Rv HGVVELAASYDHVGPITRSAHDAAVLLSVIAGSDIHD----PSCSAEPVP
MMAR_0937|M.marinum_M SGVIGVASPQDMVGPMARTVTDVATLLTVMTGVDDSD-PTTRAGGAHTAT
MUL_0689|M.ulcerans_Agy99 SGVIGVASPQDMVGPMARTVTDVATLLTVMTGVDDSD-PTTRAGGAHTAT
MSMEG_3400|M.smegmatis_MC2_155 TGFSPLVHFQDTPGPMARNVSDLAAVLDVIVGYDPTDSYTALATSGPEVG
*. . * ** *.. * * : *:.* * * : .
MLBr_01702|M.leprae_Br4923 DIVGAAKAGESGDLHGVRVGVVRQLR-GEGYQSGVLASFQAAVEQLIALG
MAV_3859|M.avium_104 DVVGAARAGAAGDLKGVRVGVVKQLR-GEGYQPGVLASFEAAVEQLTALG
TH_2639|M.thermoresistible__bu DVVAAARAGASGDLKGVRVGVVRQLR-GEGIQPGVLASFDAAVEQLAALG
Mflv_4246|M.gilvum_PYR-GCK DVVAAARTGAGGDLTGVRIGVVKQLRSGEGYQAGVLASFNAAVDQLTALG
Mvan_2115|M.vanbaalenii_PYR-1 DVVGAARAGARGDLKGVRIGVVKQLRSGEGYQPGVLASFTAAVEQLTALG
MAB_3341c|M.abscessus_ATCC_199 DVVAAARAGASGDLKGVRVGVVTQLR-GDGYQPGVMASFDAAVEQLTALG
Mb3409|M.bovis_AF2122/97 DYAADLALTRIP-----RVGVDWSQT--TSFDEDTTAMLADVVKTLDDIG
Rv3375|M.tuberculosis_H37Rv DYAADLALTRIP-----RVGVDWSQT--TSFDEDTTAMLADVVKTLDDIG
MMAR_0937|M.marinum_M DYRRFLD---PAALQGARLGVARERFGA---HEATDALIEGALGQLAALG
MUL_0689|M.ulcerans_Agy99 DYRRFLD---PAALQGARLGVARERFGA---HEATDALIEGALGQLAALG
MSMEG_3400|M.smegmatis_MC2_155 DYGEALDGVDADTLSGFRVGVLTDAFGAGDDQELTNSVVRAAIDRLRHHG
* *:** . . . : . .: * *
MLBr_01702|M.leprae_Br4923 ATVSE-VDCPHFDYALAAYYLILPSEVSSNLARFDAMRYGLRVGDDGSRS
MAV_3859|M.avium_104 AEVSE-VDCPHFEYALAAYYLILPSEVSSNLARFDAMRYGLRIGDDGSHS
TH_2639|M.thermoresistible__bu AEVIE-VDCPNIDHALAAYYLIMPSEVSSNLARFDAMRYGLRVGDDGTRS
Mflv_4246|M.gilvum_PYR-GCK AEVTE-VDCPHFDYSLPAYYLILPSEVSSNLAKFDGMRYGLRAGDDGTHS
Mvan_2115|M.vanbaalenii_PYR-1 AEVSE-VDCPHFDHSLAAYYLILPSEVSSNLAKFDGMRYGLRVGDDGTTS
MAB_3341c|M.abscessus_ATCC_199 ADVVE-VSCPHFEYALPAYYLILPSEVSSNLARFDAMRYGMRVGDDGTHS
Mb3409|M.bovis_AF2122/97 WPVID-VKLPALAPMVAAFGKMRAVET----AIAHADTYPARADEYG---
Rv3375|M.tuberculosis_H37Rv WPVID-VKLPALAPMVAAFGKMRAVET----AIAHADTYPARADEYG---
MMAR_0937|M.marinum_M AEIVDPIQASSLPFFGDLELELFRYGLKASLNGYLGAHPRAAVGSLDELI
MUL_0689|M.ulcerans_Agy99 AEIVDPIQASSLPFFGDLELELFRYGLKASLNGYLGAHPRAAVGSLDEPI
MSMEG_3400|M.smegmatis_MC2_155 TGVVDGIVLGNLETW-VAETSLYTIQSKFDLEKFLGSRRHTGPTTIREIV
: : : : : .
MLBr_01702|M.leprae_Br4923 AEEVIALTRAAGFGPEVKRRIMIGAYALSAGYYDSYYSQAQKVRTLIARD
MAV_3859|M.avium_104 AEEVMALTRAAGFGPEVKRRIMIGTYALSAGYYDAYYNQAQKVRTLIARD
TH_2639|M.thermoresistible__bu AEEVMALTRAAGFGPEVKRRIMIGTYALSAGYYDAYYNQAQKVRTLVARD
Mflv_4246|M.gilvum_PYR-GCK AEEVMALTRAAGFGPEVKRRIMIGAYALSAGYYDAYYNQAQKVRTLIARD
Mvan_2115|M.vanbaalenii_PYR-1 AEEVMAMTRAAGFGAEVKRRIMIGTYALSAGYYDAYYNQAQKVRTLIARD
MAB_3341c|M.abscessus_ATCC_199 AEEVMALTRAAGFGPEVKRRIMIGTYALSAGYYDAYYGRAQKVRTLIKRD
Mb3409|M.bovis_AF2122/97 -------------------PIMRAMIDAGHRLAAVEYQTLTERRLEFTRS
Rv3375|M.tuberculosis_H37Rv -------------------PIMRAMIDAGHRLAAVEYQTLTERRLEFTRS
MMAR_0937|M.marinum_M AFNRAHAGQVMPYFGQEFLEQSQAKGDLTDPQYLRVRAELRRLAGADGID
MUL_0689|M.ulcerans_Agy99 AFNRAHAGQVMPYFGQEFLEQSQAKGDLTDSQYLRVRAELRRLAGADGID
MSMEG_3400|M.smegmatis_MC2_155 DHGDFHP---LTDLLADIADGPSNPEDHPEYFKKRTRQEDWRRT----LL
.
MLBr_01702|M.leprae_Br4923 LDKAYRSVDVLVSPAT---PTTAFSLGEKADDPLAMYLFDLCTLPLNLAG
MAV_3859|M.avium_104 LDAAYESVDVVVSPAT---PTTAFGLGEKVDDPLAMYLFDLCTLPLNLAG
TH_2639|M.thermoresistible__bu FDAAYEKVDVLVSPTT---PTTAFPLGEKVDHPLAMYLFDLCTLPLNLAG
Mflv_4246|M.gilvum_PYR-GCK LDAAYQKVDVLVSPAT---PTTAFRLGEKVDDPLSMYLFDLCTLPLNLAG
Mvan_2115|M.vanbaalenii_PYR-1 LDEAYRSVDVLVSPAT---PSTAFRLGEKVDDPLAMYLFDLCTLPLNLAG
MAB_3341c|M.abscessus_ATCC_199 LERAYEQVDVLVTPAT---PTTAFRLGEKVDDPLAMYQFDLCTLPLNLAG
Mb3409|M.bovis_AF2122/97 LRRVFHDVDILLMPSAGIASPTLETMRGLGQDPELTARLAMPTAPFNVSG
Rv3375|M.tuberculosis_H37Rv LRRVFHDVDILLMPSAGIASPTLETMRGLGQDPELTARLAMPTAPFNVSG
MMAR_0937|M.marinum_M KALREHRLDAIVAPTE---GSPAFAIDPVVGDNILPGGCSTPPAVAG---
MUL_0689|M.ulcerans_Agy99 KALREHRLDAIVAPTE---GSPAFAIDPVVGDNILPGGCSTPPAVAG---
MSMEG_3400|M.smegmatis_MC2_155 AKMADAEIDFLVYPTV---QVPAPTRADLAAKRWTALNFPTNTVIASQTS
:* :: *: . . . .
MLBr_01702|M.leprae_Br4923 HCGMSVPSGLSSDDDLPVGLQIMAPALADDRLYRVGAAYEAARGPLPSAI
MAV_3859|M.avium_104 HCGMSVPSGLSPDDDLPVGLQIMAPALADDRLYRVGAAYEAARGPLRSAI
TH_2639|M.thermoresistible__bu HCGMSVPSGLSPDDNLPVGLQIMAPALADDRLYRVGAAYEAARGPLPAAV
Mflv_4246|M.gilvum_PYR-GCK HCGMSVPSGLSADDNLPVGLQIMAPALADDRLYRVGAAYEAARGPLPTAL
Mvan_2115|M.vanbaalenii_PYR-1 HCGMSVPSGLSADDNLPVGLQIMAPALADDRLYRVGAAYEAARGPLPTAL
MAB_3341c|M.abscessus_ATCC_199 HCGMSVPSGLSADDGLPVGLQIMAPALADDRLYRVGAAYETARGALPAAL
Mb3409|M.bovis_AF2122/97 NPAICLPAGTTARG-TPLGVQFIGREFDEHLLVRAGHAFQQVTGYHRRRP
Rv3375|M.tuberculosis_H37Rv NPAICLPAGTTARG-TPLGVQFIGREFDEHLLVRAGHAFQQVTGYHRRRP
MMAR_0937|M.marinum_M YPHICVPAGYFCG--LPVGLSLFAGAFQEPKLIGYAYAFEQATGVRRPPR
MUL_0689|M.ulcerans_Agy99 YPHICVPAGYFCG--LPVGLSLFAGAFQEPKLIGYAYAFEQATGVRRPPR
MSMEG_3400|M.smegmatis_MC2_155 LPAMSIPVGFTDSG-LPVGLEVVGRPLTERALLRFARAWELAEAPRRQPQ
:.:* * *:*:.... : : * . *:: . .
MLBr_01702|M.leprae_Br4923 ----------
MAV_3859|M.avium_104 ----------
TH_2639|M.thermoresistible__bu ----------
Mflv_4246|M.gilvum_PYR-GCK ----------
Mvan_2115|M.vanbaalenii_PYR-1 ----------
MAB_3341c|M.abscessus_ATCC_199 ----------
Mb3409|M.bovis_AF2122/97 PV--------
Rv3375|M.tuberculosis_H37Rv PV--------
MMAR_0937|M.marinum_M FVPTLST---
MUL_0689|M.ulcerans_Agy99 FVPTLST---
MSMEG_3400|M.smegmatis_MC2_155 IEASKARVTM