For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MELYELPLIEVAEKIRTKEVSPVEVAESSLARLEEVEPLLTAFVTTTPELALEQAKAAEKEIADGKYRGP LHGIPLGVKDLYDTAGIRTTSSSAQRADYVPDADSVSVAKLYDAGMVLVGKTHTHEFAYGATTPTTGNPW APDRTPGGSSGGSGAAVAAGVVHVALGSDTGGSIRIPAALCGTVGLKPTYGRASRVGVASLSWSLDHVGP LSRNVTDAALVMQAMSGYDRRDPGTANVAVPDMVSGIDAGVAGKKIGIPVNYYTDRVAPEAAEAAKVAAA TFEKLGAQLVEVEIPMAEHIVPTEWAIMMPEATAYHMDYLRNSPEKFTDEVRTLLEVGAAEPAVDYVNAM RLRTLIQAAWNEMFTGIDVLLAPTVVAPATLRSDPFVRWEDGTVEAATAAYVRLSAPANVTGLPSLSVPA AFTADGLPLGVQILGKPFAEPEILTFGYALEQNTDTVGRIAPVLEKVG
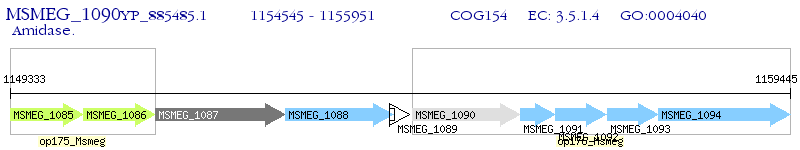
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1090 | - | - | 100% (468) | amidase |
| M. smegmatis MC2 155 | MSMEG_1088 | gatA | 4e-95 | 41.83% (447) | glutamyl-tRNA(Gln)/aspartyl-tRNA(Asn) amidotransferase, A |
| M. smegmatis MC2 155 | MSMEG_2986 | - | 8e-61 | 36.50% (452) | amidohydrolase, AtzE family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3409 | amiD | 3e-73 | 35.85% (463) | amidase AmiD |
| M. gilvum PYR-GCK | Mflv_4246 | gatA | 3e-60 | 32.38% (488) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. tuberculosis H37Rv | Rv3375 | amiD | 3e-73 | 35.85% (463) | amidase AmiD |
| M. leprae Br4923 | MLBr_01702 | gatA | 1e-59 | 32.99% (488) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. abscessus ATCC 19977 | MAB_3341c | gatA | 1e-62 | 34.17% (477) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. marinum M | MMAR_0984 | amiD | 6e-71 | 34.57% (460) | amidase, AmiD |
| M. avium 104 | MAV_3859 | gatA | 7e-57 | 32.43% (478) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. thermoresistible (build 8) | TH_2639 | - | 1e-58 | 31.21% (487) | PUTATIVE glutamyl-tRNA (Gln) amidotransferase (subunit A), GatA |
| M. ulcerans Agy99 | MUL_1939 | gatA | 7e-60 | 32.85% (478) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. vanbaalenii PYR-1 | Mvan_2115 | gatA | 1e-59 | 32.93% (492) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_01702|M.leprae_Br4923 ---------MIGSLHEIIRLDASTLAAKIVAKELSSVEITQACLDQIEAT
MAV_3859|M.avium_104 -------------MNEIIRSDAATLAARIAAKELSSVEATQACLDQIAAT
MUL_1939|M.ulcerans_Agy99 -------------MTDLIRSDAATLAAKIAAKEVSATELTQACLDQIEAT
TH_2639|M.thermoresistible__bu -------MSQGKSTDDLIRLDAATLGAKIAAKEISSTELTQACLDRIAAT
Mflv_4246|M.gilvum_PYR-GCK -------------MSELIKLDAATLADKISSKEVSSAEVTQAFLDQIAAT
Mvan_2115|M.vanbaalenii_PYR-1 -------------MTDLIKLDAATLAAKIAAREVSATEVTQACLDQIAAT
MAB_3341c|M.abscessus_ATCC_199 -------------MTDLIRQDAAALAALIHSGEVSSVEVTRAHLDQIAST
Mb3409|M.bovis_AF2122/97 MTDADSAVPPRLDEDAISKLELTEVADLIRTRQLTSAEVTESTLRRIERL
Rv3375|M.tuberculosis_H37Rv MTDADSAVPPRLDEDAISKLELTEVADLIRTRQLTSAEVTESTLRRIERL
MMAR_0984|M.marinum_M MSDIDPCYAT-----------IAELSHAFATRALSPVEVTRAHLQRIDRL
MSMEG_1090|M.smegmatis_MC2_155 ---MELYELP-----------LIEVAEKIRTKEVSPVEVAESSLARLEEV
:. : : ::..* :.: * ::
MLBr_01702|M.leprae_Br4923 DDTYRAFLHVAAEKALSAAAAVDK----AVAAGGQLSSTLAGVPLALKDV
MAV_3859|M.avium_104 DERYHAFLHVAADEALGAAAVVDE----AVAAGERLPSPLAGVPLALKDV
MUL_1939|M.ulcerans_Agy99 DDRYHAFLHIGAHEALSAAAAVDT----ALAAGERLPSALAGVPLALKDV
TH_2639|M.thermoresistible__bu DDRYNAFLHVAANKALSAAARVDA----AVAAGEELPSPLAGVPLALKDV
Mflv_4246|M.gilvum_PYR-GCK DGDYHAFLHVGAEQALAAAQTVDR----AVAAGEHLPSPLAGVPLALKDV
Mvan_2115|M.vanbaalenii_PYR-1 DAEYHAFLHVAGDQALAAAATVDKAIHEATAAGERLPSPLAGVPLALKDV
MAB_3341c|M.abscessus_ATCC_199 DETYRAFLHVAGEQALAAAKDVDD----AIAAGS-VPSGLAGVPLALKDV
Mb3409|M.bovis_AF2122/97 DPQLKSYAFVMPETALAAARAADA-----DIARGHYEGVLHGVPIGVKDL
Rv3375|M.tuberculosis_H37Rv DPQLKSYAFVMPETALAAARAADA-----DIARGHYEGVLHGVPIGVKDL
MMAR_0984|M.marinum_M NPVLHPYMTVLADSALDEARQAEE-----QIARGNHRGRLHGVPIAVKDL
MSMEG_1090|M.smegmatis_MC2_155 EPLLTAFVTTTPELALEQAKAAEK-----EIADGKYRGPLHGIPLGVKDL
: .: . ** * .: * . * *:*:.:**:
MLBr_01702|M.leprae_Br4923 FTTVDMPTTCGSKILQGWHSPYDATVTTRLRAAGIPILGKTNMDEFAMGS
MAV_3859|M.avium_104 FTTVDMPTTCGSKILQGWRSPYDATVTTKLRAAGIPILGKTNMDEFAMGS
MUL_1939|M.ulcerans_Agy99 FTTVDMPTTCGSKILEGWRSPYDATLTLRLRAAGIPILGKTNMDEFAMGS
TH_2639|M.thermoresistible__bu FTTIDMPTTCGSKILDGWQAPYDATVTVALRAAGIPILGKTNLDEFAMGS
Mflv_4246|M.gilvum_PYR-GCK FTTTDMPTTCGSKVLEGWTSPYDATVTAKLRSAGIPILGKTNMDEFAMGS
Mvan_2115|M.vanbaalenii_PYR-1 FTTTDMPTTCGSKILEGWTSPYDATVTAKLRAAGIPILGKTNMDEFAMGS
MAB_3341c|M.abscessus_ATCC_199 FTTRDMPTTCGSKILENWVPPYDATVTARLRGAGIPILGKTNMDEFAMGS
Mb3409|M.bovis_AF2122/97 CYTVDAPTAAGTTIFRDFRPAYDATVVARLRAAGAVIIGKLAMTEGAYLG
Rv3375|M.tuberculosis_H37Rv CYTVDAPTAAGTTIFRDFRPAYDATVVARLRAAGAVIIGKLAMTEGAYLG
MMAR_0984|M.marinum_M CFTQGIPTTAGMAIHRDFRPSYDATVVSRLRRAGAVLLGKLHMCEAAGAE
MSMEG_1090|M.smegmatis_MC2_155 YDTAGIRTTSSSAQRADYVPDADSVSVAKLYDAGMVLVGKTHTHEFAYG-
* . *:.. .: . *:. . * ** ::** * *
MLBr_01702|M.leprae_Br4923 STENSAYGPTRNPWNVDRVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQ
MAV_3859|M.avium_104 STENSAYGPTRNPWNVERVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQ
MUL_1939|M.ulcerans_Agy99 STENSAYGPTRNPWNLDRVPGGSGGGSAAALAAYQAPLAIGSDTGGSIRQ
TH_2639|M.thermoresistible__bu STENSAFGPTRNPWDVNRVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQ
Mflv_4246|M.gilvum_PYR-GCK STENSAYGPTRNPWDTERVPGGSGGGSAAALAAFQAPLAIGTDTGGSIRQ
Mvan_2115|M.vanbaalenii_PYR-1 STENSAYGPTRNPWNVDCVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQ
MAB_3341c|M.abscessus_ATCC_199 STENSAYGPTRNPWDVERVPGGSGGGSAAALAAFQAPLAIGTDTGGSIRQ
Mb3409|M.bovis_AF2122/97 YHPSLPT--PVNPWDPTAWAGVSSSGCGVATAAGLCFGSIGSDTGGSIRF
Rv3375|M.tuberculosis_H37Rv YHPSLPT--PVNPWDPTAWAGVSSSGCGVATAAGLCFGSIGSDTGGSIRF
MMAR_0984|M.marinum_M YHPDFPA--VVNPWSAERWAGASSSGSAVATVTGLCTASLGSDTGGSIRT
MSMEG_1090|M.smegmatis_MC2_155 --ATTPT--TGNPWAPDRTPGGSSGGSGAAVAAGVVHVALGSDTGGSIRI
. *** .* *..*...* .: ::*:*******
MLBr_01702|M.leprae_Br4923 PAALTATVGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTALLHAVIA
MAV_3859|M.avium_104 PAALTATVGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTALLHQVIA
MUL_1939|M.ulcerans_Agy99 PAALTATVGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTAMLHQVIA
TH_2639|M.thermoresistible__bu PAALTATVGVKPTYGTVSRYGLVACASSLDQGGPCGRTVLDTALLHQVIA
Mflv_4246|M.gilvum_PYR-GCK PAALTATVGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTALLHQVIA
Mvan_2115|M.vanbaalenii_PYR-1 PAALTATVGVKPTYGTVSRYGLIACASSLDQGGPCARTVLDTALLHQVIA
MAB_3341c|M.abscessus_ATCC_199 PAALTATVGVKPTYGTVSRFGLVACASSLDQGGPCARTVLDTALLHQVIA
Mb3409|M.bovis_AF2122/97 PTSMCGVTGIKPTWGRVSRHGVVELAASYDHVGPITRSAHDAAVLLSVIA
Rv3375|M.tuberculosis_H37Rv PTSMCGVTGIKPTWGRVSRHGVVELAASYDHVGPITRSAHDAAVLLSVIA
MMAR_0984|M.marinum_M PCTMTGATGLKPTWGRVSVHGVFAMAPSLDTIGPMARNAEDTGHVLRAIA
MSMEG_1090|M.smegmatis_MC2_155 PAALCGTVGLKPTYGRASRVGVASLSWSLDHVGPLSRNVTDAALVMQAMS
* :: ...*:***:* .* *: : * * ** *.. *:. : .::
MLBr_01702|M.leprae_Br4923 GHDARDSTSVEAAVPDIVGAAKAGESGDLHGVRVGVVRQLR-GEGYQSGV
MAV_3859|M.avium_104 GHDIRDSTSVDAPVPDVVGAARAGAAGDLKGVRVGVVKQLR-GEGYQPGV
MUL_1939|M.ulcerans_Agy99 GHDAKDSTSLETEIPDVVGAAKAGASGDLRGVRIGVVKQLR-GDGYQPGV
TH_2639|M.thermoresistible__bu GHDAKDSTSLTAPVPDVVAAARAGASGDLKGVRVGVVRQLR-GEGIQPGV
Mflv_4246|M.gilvum_PYR-GCK GHDPLDSTSVEAPVPDVVAAARTGAGGDLTGVRIGVVKQLRSGEGYQAGV
Mvan_2115|M.vanbaalenii_PYR-1 GHDPRDSTSVNAAVPDVVGAARAGARGDLKGVRIGVVKQLRSGEGYQPGV
MAB_3341c|M.abscessus_ATCC_199 GHDPRDSTSVDEPVPDVVAAARAGASGDLKGVRVGVVTQLR-GDGYQPGV
Mb3409|M.bovis_AF2122/97 GSDIHDPSCSAEPVPDYAADLAL-----TRIPRVG-VDWSQTTS-FDEDT
Rv3375|M.tuberculosis_H37Rv GSDIHDPSCSAEPVPDYAADLAL-----TRIPRVG-VDWSQTTS-FDEDT
MMAR_0984|M.marinum_M GADPQDPTATRVGVPEYVTGSPE-----VRTVRIG-FDPEYALGGVEPDV
MSMEG_1090|M.smegmatis_MC2_155 GYDRRDPGTANVAVPDMVSGIDAG----VAGKKIG-IPVNYYTDRVAPEA
* * *. :*: . ::* . .
MLBr_01702|M.leprae_Br4923 LASFQAAVEQLIALGATVSEVDCPHFDYALAAYYLILPSEVSSNLARFDA
MAV_3859|M.avium_104 LASFEAAVEQLTALGAEVSEVDCPHFEYALAAYYLILPSEVSSNLARFDA
MUL_1939|M.ulcerans_Agy99 LASFEAAVAQLTALGAEVSEVDCPHFDHALAAYYLILPSEVSSNLARFDA
TH_2639|M.thermoresistible__bu LASFDAAVEQLAALGAEVIEVDCPNIDHALAAYYLIMPSEVSSNLARFDA
Mflv_4246|M.gilvum_PYR-GCK LASFNAAVDQLTALGAEVTEVDCPHFDYSLPAYYLILPSEVSSNLAKFDG
Mvan_2115|M.vanbaalenii_PYR-1 LASFTAAVEQLTALGAEVSEVDCPHFDHSLAAYYLILPSEVSSNLAKFDG
MAB_3341c|M.abscessus_ATCC_199 MASFDAAVEQLTALGADVVEVSCPHFEYALPAYYLILPSEVSSNLARFDA
Mb3409|M.bovis_AF2122/97 TAMLADVVKTLDDIGWPVIDVKLPALAPMVAAFGKMRAVETAIAHADTYP
Rv3375|M.tuberculosis_H37Rv TAMLADVVKTLDDIGWPVIDVKLPALAPMVAAFGKMRAVETAIAHADTYP
MMAR_0984|M.marinum_M ARVVRTALRVLADLGADIREVSLPSTAALAAGWGAYVGAQIAVVHERTYP
MSMEG_1090|M.smegmatis_MC2_155 AEAAKVAAATFEKLGAQLVEVEIPMAEHIVPTEWAIMMPEATAYHMDYLR
. : :* : :*. * . : :
MLBr_01702|M.leprae_Br4923 MRYGLRVGDDGSRSAEEVIALTRAAGFGPEVKRRIMIGAYALSAGYYDSY
MAV_3859|M.avium_104 MRYGLRIGDDGSHSAEEVMALTRAAGFGPEVKRRIMIGTYALSAGYYDAY
MUL_1939|M.ulcerans_Agy99 MRYGLRIGDDGTHSAEEVMAMTRAAGFGPEVKRRIMIGAYALSAGYYDAY
TH_2639|M.thermoresistible__bu MRYGLRVGDDGTRSAEEVMALTRAAGFGPEVKRRIMIGTYALSAGYYDAY
Mflv_4246|M.gilvum_PYR-GCK MRYGLRAGDDGTHSAEEVMALTRAAGFGPEVKRRIMIGAYALSAGYYDAY
Mvan_2115|M.vanbaalenii_PYR-1 MRYGLRVGDDGTTSAEEVMAMTRAAGFGAEVKRRIMIGTYALSAGYYDAY
MAB_3341c|M.abscessus_ATCC_199 MRYGMRVGDDGTHSAEEVMALTRAAGFGPEVKRRIMIGTYALSAGYYDAY
Mb3409|M.bovis_AF2122/97 A---------------------RADEYGPIMRAMIDAG-----HRLAAVE
Rv3375|M.tuberculosis_H37Rv A---------------------RADEYGPIMRAMIDAG-----HRLAAVE
MMAR_0984|M.marinum_M S---------------------RSAEYGPFASQLLDAG-----HAVSGME
MSMEG_1090|M.smegmatis_MC2_155 N---------------------SPEKFTDEVRTLLEVG-----AAEPAVD
. : : *
MLBr_01702|M.leprae_Br4923 YSQAQKVRTLIARDLDKAYRSVDVLVSP------ATPTTAFSLGEKADDP
MAV_3859|M.avium_104 YNQAQKVRTLIARDLDAAYESVDVVVSP------ATPTTAFGLGEKVDDP
MUL_1939|M.ulcerans_Agy99 YNQAQKIRTLIARDLDEAYQSVDVLVSP------ATPTTAFPLGEKVDDP
TH_2639|M.thermoresistible__bu YNQAQKVRTLVARDFDAAYEKVDVLVSP------TTPTTAFPLGEKVDHP
Mflv_4246|M.gilvum_PYR-GCK YNQAQKVRTLIARDLDAAYQKVDVLVSP------ATPTTAFRLGEKVDDP
Mvan_2115|M.vanbaalenii_PYR-1 YNQAQKVRTLIARDLDEAYRSVDVLVSP------ATPSTAFRLGEKVDDP
MAB_3341c|M.abscessus_ATCC_199 YGRAQKVRTLIKRDLERAYEQVDVLVTP------ATPTTAFRLGEKVDDP
Mb3409|M.bovis_AF2122/97 YQTLTERRLEFTRSLRRVFHDVDILLMP-SAGIASPTLETMR--GLGQDP
Rv3375|M.tuberculosis_H37Rv YQTLTERRLEFTRSLRRVFHDVDILLMP-SAGIASPTLETMR--GLGQDP
MMAR_0984|M.marinum_M IARIQRERNLFAGRLAALFEDVDLLVAP-VLPLANLTLDRFR--ELLFNP
MSMEG_1090|M.smegmatis_MC2_155 YVNAMRLRTLIQAAWNEMFTGIDVLLAPTVVAPATLRSDPFVRWEDGTVE
. * . : :*::: * :
MLBr_01702|M.leprae_Br4923 LAMYLFDLCTLPLNLAGHCGMSVPSGLSSDDDLPVGLQIMAPALADDRLY
MAV_3859|M.avium_104 LAMYLFDLCTLPLNLAGHCGMSVPSGLSPDDDLPVGLQIMAPALADDRLY
MUL_1939|M.ulcerans_Agy99 LAMYLFDLCTLPLNLAGHCGMSVPSGLSPDDGLPVGLQIMAPALADDRLY
TH_2639|M.thermoresistible__bu LAMYLFDLCTLPLNLAGHCGMSVPSGLSPDDNLPVGLQIMAPALADDRLY
Mflv_4246|M.gilvum_PYR-GCK LSMYLFDLCTLPLNLAGHCGMSVPSGLSADDNLPVGLQIMAPALADDRLY
Mvan_2115|M.vanbaalenii_PYR-1 LAMYLFDLCTLPLNLAGHCGMSVPSGLSADDNLPVGLQIMAPALADDRLY
MAB_3341c|M.abscessus_ATCC_199 LAMYQFDLCTLPLNLAGHCGMSVPSGLSADDGLPVGLQIMAPALADDRLY
Mb3409|M.bovis_AF2122/97 ELTARLAMPTAPFNVSGNPAICLPAGTTARG-TPLGVQFIGREFDEHLLV
Rv3375|M.tuberculosis_H37Rv ELTARLAMPTAPFNVSGNPAICLPAGTTARG-TPLGVQFIGREFDEHLLV
MMAR_0984|M.marinum_M ETLPDLLRFTGPFNFSGSPTITLPGGFDSHG-GPIGFQFVARHFDEPLLI
MSMEG_1090|M.smegmatis_MC2_155 AATAAYVRLSAPANVTGLPSLSVPAAFTADG-LPLGVQILGKPFAEPEIL
: * *.:* : :*.. . . *:*.*::. : : :
MLBr_01702|M.leprae_Br4923 RVGAAYEAARGPLPSAI-------
MAV_3859|M.avium_104 RVGAAYEAARGPLRSAI-------
MUL_1939|M.ulcerans_Agy99 RVGAAYEAARGPLPSAI-------
TH_2639|M.thermoresistible__bu RVGAAYEAARGPLPAAV-------
Mflv_4246|M.gilvum_PYR-GCK RVGAAYEAARGPLPTAL-------
Mvan_2115|M.vanbaalenii_PYR-1 RVGAAYEAARGPLPTAL-------
MAB_3341c|M.abscessus_ATCC_199 RVGAAYETARGALPAAL-------
Mb3409|M.bovis_AF2122/97 RAGHAFQQVTGYHRRRPPV-----
Rv3375|M.tuberculosis_H37Rv RAGHAFQQVTGYHRRRPPV-----
MMAR_0984|M.marinum_M RAGRAFQSATDWHLRRPPVQ----
MSMEG_1090|M.smegmatis_MC2_155 TFGYALEQNTDTVGRIAPVLEKVG
* * : .