For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MELPEFTIAETQTAFERGEWTAAGLTDCYLRRIREIDQSGPMLRSIIEVNSDALAIAEALDAERSGGRIR GALHGVPVVIKDSIDTGDKMATTAGSLALEGNIATRDAFVVKQLRDAGAVILGKANMSEWGNMSEWGYMR STRPCSGWSSRGGQVRNPYVLDRSPLGSSSSSAVAVAANLCVAALGAEVDGSIVRPASSNSIVGLKPTVG LLSRSGVIGVASPQDMVGPMARTVTDVATLLTVMTGVDDSDPTTRAGGAHTATDYRRFLDPAALQGARLG VARERFGAHEATDALIEGALGQLAALGAEIVDPIQASSLPFFGDLELELFRYGLKASLNGYLGAHPRAAV GSLDEPIAFNRAHAGQVMPYFGQEFLEQSQAKGDLTDSQYLRVRAELRRLAGADGIDKALREHRLDAIVA PTEGSPAFAIDPVVGDNILPGGCSTPPAVAGYPHICVPAGYFCGLPVGLSLFAGAFQEPKLIGYAYAFEQ ATGVRRPPRFVPTLST
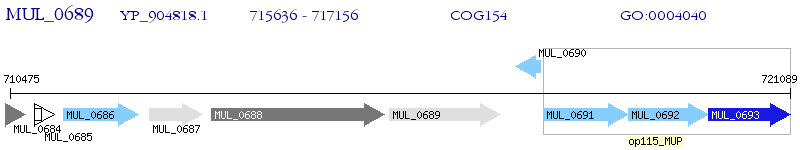
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0689 | gatA_1 | - | 100% (506) | peptide amidase, GatA_1 |
| M. ulcerans Agy99 | MUL_1939 | gatA | 1e-43 | 32.94% (504) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. ulcerans Agy99 | MUL_3616 | amiA2 | 8e-26 | 28.63% (503) | amidase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3036c | gatA | 4e-45 | 33.33% (486) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. gilvum PYR-GCK | Mflv_4246 | gatA | 4e-43 | 32.43% (518) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. tuberculosis H37Rv | Rv3011c | gatA | 1e-45 | 33.27% (502) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. leprae Br4923 | MLBr_01702 | gatA | 2e-44 | 32.41% (506) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. abscessus ATCC 19977 | MAB_3341c | gatA | 3e-43 | 32.07% (502) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. marinum M | MMAR_0937 | gatA_1 | 0.0 | 98.02% (506) | peptide amidase, GatA_1 |
| M. avium 104 | MAV_3859 | gatA | 1e-40 | 32.77% (470) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. smegmatis MC2 155 | MSMEG_3400 | - | 5e-52 | 32.63% (524) | glutamyl-tRNA(Gln) amidotransferase subunit A |
| M. thermoresistible (build 8) | TH_2639 | - | 3e-40 | 31.74% (501) | PUTATIVE glutamyl-tRNA (Gln) amidotransferase (subunit A), GatA |
| M. vanbaalenii PYR-1 | Mvan_2115 | gatA | 1e-41 | 32.87% (505) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_01702|M.leprae_Br4923 --------------------------------------------------
MAV_3859|M.avium_104 --------------------------------------------------
Mb3036c|M.bovis_AF2122/97 --------------------------------------------------
Rv3011c|M.tuberculosis_H37Rv --------------------------------------------------
TH_2639|M.thermoresistible__bu --------------------------------------------------
Mflv_4246|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2115|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3341c|M.abscessus_ATCC_199 --------------------------------------------------
MUL_0689|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0937|M.marinum_M --------------------------------------------------
MSMEG_3400|M.smegmatis_MC2_155 MVSHSKDSAETPTLAESVYQRIRRNILEGRIMPGTRVSIRSLAESVGVST
MLBr_01702|M.leprae_Br4923 --------------------------------------------------
MAV_3859|M.avium_104 --------------------------------------------------
Mb3036c|M.bovis_AF2122/97 --------------------------------------------------
Rv3011c|M.tuberculosis_H37Rv --------------------------------------------------
TH_2639|M.thermoresistible__bu --------------------------------------------------
Mflv_4246|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2115|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3341c|M.abscessus_ATCC_199 --------------------------------------------------
MUL_0689|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0937|M.marinum_M --------------------------------------------------
MSMEG_3400|M.smegmatis_MC2_155 MPTREALKRLSFEGLVEYDRRAVTISTLSPKAIRELFTIRLRLELLATEW
MLBr_01702|M.leprae_Br4923 --------------------------------------------------
MAV_3859|M.avium_104 --------------------------------------------------
Mb3036c|M.bovis_AF2122/97 --------------------------------------------------
Rv3011c|M.tuberculosis_H37Rv --------------------------------------------------
TH_2639|M.thermoresistible__bu --------------------------------------------------
Mflv_4246|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2115|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3341c|M.abscessus_ATCC_199 --------------------------------------------------
MUL_0689|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0937|M.marinum_M --------------------------------------------------
MSMEG_3400|M.smegmatis_MC2_155 ALPRLDAETTVALRSVLDRMIVPGISAITWRELNREFHQTFYSCGDSRYL
MLBr_01702|M.leprae_Br4923 --------------------------------------------------
MAV_3859|M.avium_104 --------------------------------------------------
Mb3036c|M.bovis_AF2122/97 --------------------------------------------------
Rv3011c|M.tuberculosis_H37Rv --------------------------------------------------
TH_2639|M.thermoresistible__bu --------------------------------------------------
Mflv_4246|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2115|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3341c|M.abscessus_ATCC_199 --------------------------------------------------
MUL_0689|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0937|M.marinum_M --------------------------------------------------
MSMEG_3400|M.smegmatis_MC2_155 LELIHNIWDRIQPYMAIYASAMHDFTEADRQHEHMYQLMLARDLDGLLDA
MLBr_01702|M.leprae_Br4923 ----------------------MIGSLHEIIRLDASTLAAKIVAKELSSV
MAV_3859|M.avium_104 --------------------------MNEIIRSDAATLAARIAAKELSSV
Mb3036c|M.bovis_AF2122/97 --------------------------MTDIIRSDAATLAAKIAIKEVSST
Rv3011c|M.tuberculosis_H37Rv --------------------------MTDIIRSDAATLAAKIAIKEVSSA
TH_2639|M.thermoresistible__bu --------------------MSQGKSTDDLIRLDAATLGAKIAAKEISST
Mflv_4246|M.gilvum_PYR-GCK --------------------------MSELIKLDAATLADKISSKEVSSA
Mvan_2115|M.vanbaalenii_PYR-1 --------------------------MTDLIKLDAATLAAKIAAREVSAT
MAB_3341c|M.abscessus_ATCC_199 --------------------------MTDLIRQDAAALAALIHSGEVSSV
MUL_0689|M.ulcerans_Agy99 ---------------------------MELPEFTIAETQTAFERGEWTAA
MMAR_0937|M.marinum_M ---------------------------MELPEFTIAETQTAFERGEWTAA
MSMEG_3400|M.smegmatis_MC2_155 TTRHLEHTAEAIVSALGADTTTGRGNNVNYEELSISSFHRLVESGGATSA
: . : . ::.
MLBr_01702|M.leprae_Br4923 EITQACLDQIEATD------DTYRAFLHVAAEKALSAAAAVDK----AVA
MAV_3859|M.avium_104 EATQACLDQIAATD------ERYHAFLHVAADEALGAAAVVDE----AVA
Mb3036c|M.bovis_AF2122/97 EITRACLDQIEATD------ETYHAFLHVAADEALAAAAAVDK----QVA
Rv3011c|M.tuberculosis_H37Rv EITRACLDQIEATD------ETYHAFLHVAADEALAAAAAIDK----QVA
TH_2639|M.thermoresistible__bu ELTQACLDRIAATD------DRYNAFLHVAANKALSAAARVDA----AVA
Mflv_4246|M.gilvum_PYR-GCK EVTQAFLDQIAATD------GDYHAFLHVGAEQALAAAQTVDR----AVA
Mvan_2115|M.vanbaalenii_PYR-1 EVTQACLDQIAATD------AEYHAFLHVAGDQALAAAATVDKAIHEATA
MAB_3341c|M.abscessus_ATCC_199 EVTRAHLDQIASTD------ETYRAFLHVAGEQALAAAKDVDD----AIA
MUL_0689|M.ulcerans_Agy99 GLTDCYLRRIREID----QSGPMLRSIIEVNSDALAIAEALDA----ERS
MMAR_0937|M.marinum_M GLTDCYLRRIREID----QSGPMLRSIIEVNPDALAIAEALDA----ERS
MSMEG_3400|M.smegmatis_MC2_155 DLTAWYLDRIATLDSIDNPDGPQLNSVVTVNPAALDEARALDE----KYA
* * :* * : ** * :* :
MLBr_01702|M.leprae_Br4923 AGGQLSSTLAGVPLALKDVFTT-VDMPTTCGSKILQGWHSPYDATVTTRL
MAV_3859|M.avium_104 AGERLPSPLAGVPLALKDVFTT-VDMPTTCGSKILQGWRSPYDATVTTKL
Mb3036c|M.bovis_AF2122/97 AGEPLPSALAGVPLALKDVFTT-SDMPTTCGSKILEGWRSPYDATLTARL
Rv3011c|M.tuberculosis_H37Rv AGEPLPSALAGVPLALKDVFTT-SDMPTTCGSKILEGWRSPYDATLTARL
TH_2639|M.thermoresistible__bu AGEELPSPLAGVPLALKDVFTT-IDMPTTCGSKILDGWQAPYDATVTVAL
Mflv_4246|M.gilvum_PYR-GCK AGEHLPSPLAGVPLALKDVFTT-TDMPTTCGSKVLEGWTSPYDATVTAKL
Mvan_2115|M.vanbaalenii_PYR-1 AGERLPSPLAGVPLALKDVFTT-TDMPTTCGSKILEGWTSPYDATVTAKL
MAB_3341c|M.abscessus_ATCC_199 AGS-VPSGLAGVPLALKDVFTT-RDMPTTCGSKILENWVPPYDATVTARL
MUL_0689|M.ulcerans_Agy99 -GGRIRGALHGVPVVIKDSIDTGDKMATTAGSLALEGNIATRDAFVVKQL
MMAR_0937|M.marinum_M -GGRIRGALHGVPVVIKDSIDTGDKMATTAGSLALEGNIATRDAFVVKQL
MSMEG_3400|M.smegmatis_MC2_155 RTGTLVGPLHGVPILIKDQGET-KGIPTAFGSTAFADYIPDTDATVVDRL
: . * ***: :** * :.*: ** : . . ** :. *
MLBr_01702|M.leprae_Br4923 RAAGIPILGKTNMDEFAMG-----------SSTENSAYGPTRNPWNVDRV
MAV_3859|M.avium_104 RAAGIPILGKTNMDEFAMG-----------SSTENSAYGPTRNPWNVERV
Mb3036c|M.bovis_AF2122/97 RAAGIPILGKTNMDEFAMG-----------SSTENSAYGPTRNPWNLDRV
Rv3011c|M.tuberculosis_H37Rv RAAGIPILGKTNMDEFAMG-----------SSTENSAYGPTRNPWNLDRV
TH_2639|M.thermoresistible__bu RAAGIPILGKTNLDEFAMG-----------SSTENSAFGPTRNPWDVNRV
Mflv_4246|M.gilvum_PYR-GCK RSAGIPILGKTNMDEFAMG-----------SSTENSAYGPTRNPWDTERV
Mvan_2115|M.vanbaalenii_PYR-1 RAAGIPILGKTNMDEFAMG-----------SSTENSAYGPTRNPWNVDCV
MAB_3341c|M.abscessus_ATCC_199 RGAGIPILGKTNMDEFAMG-----------SSTENSAYGPTRNPWDVERV
MUL_0689|M.ulcerans_Agy99 RDAGAVILGKANMSEWGNMSEWGYMRSTRPCSGWSSRGGQVRNPYVLDRS
MMAR_0937|M.marinum_M RDAGAVILGKANMSEWG------YMRSTRPCSGWSSRGGQVRNPYVLDRS
MSMEG_3400|M.smegmatis_MC2_155 RKAGAVILGKTAMCDFAAG-----------WFSFSSRTDHTKNPYDLDRE
* ** ****: : ::. .* . .:**: :
MLBr_01702|M.leprae_Br4923 PGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTATVGVKPTYGTVSR
MAV_3859|M.avium_104 PGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTATVGVKPTYGTVSR
Mb3036c|M.bovis_AF2122/97 PGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTATVGVKPTYGTVSR
Rv3011c|M.tuberculosis_H37Rv PGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTATVGVKPTYGTVSR
TH_2639|M.thermoresistible__bu PGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTATVGVKPTYGTVSR
Mflv_4246|M.gilvum_PYR-GCK PGGSGGGSAAALAAFQAPLAIGTDTGGSIRQPAALTATVGVKPTYGTVSR
Mvan_2115|M.vanbaalenii_PYR-1 PGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTATVGVKPTYGTVSR
MAB_3341c|M.abscessus_ATCC_199 PGGSGGGSAAALAAFQAPLAIGTDTGGSIRQPAALTATVGVKPTYGTVSR
MUL_0689|M.ulcerans_Agy99 PLGSSSSSAVAVAANLCVAALGAEVDGSIVRPASSNSIVGLKPTVGLLSR
MMAR_0937|M.marinum_M PLGSSSGSAVAVAANLCVAALGAEVDGSIVRPASSNSIVGLKPTVGLLSR
MSMEG_3400|M.smegmatis_MC2_155 TGGSSAGTAAAVTANLCLVGIGEDTGGSIRLPSSFTNLFGLRVTTGLVSR
. **...:*.*::* . .:* :..*** *:: . .*:: * * :**
MLBr_01702|M.leprae_Br4923 YGLVACASSLDQGGPCARTVLDTALLHAVIAGHDARDSTSVEAAVPDIVG
MAV_3859|M.avium_104 YGLVACASSLDQGGPCARTVLDTALLHQVIAGHDIRDSTSVDAPVPDVVG
Mb3036c|M.bovis_AF2122/97 YGLVACASSLDQGGPCARTVLDTALLHQVIAGHDPRDSTSVDAEVPDVVG
Rv3011c|M.tuberculosis_H37Rv YGLVACASSLDQGGPCARTVLDTALLHQVIAGHDPRDSTSVDAEVPDVVG
TH_2639|M.thermoresistible__bu YGLVACASSLDQGGPCGRTVLDTALLHQVIAGHDAKDSTSLTAPVPDVVA
Mflv_4246|M.gilvum_PYR-GCK YGLVACASSLDQGGPCARTVLDTALLHQVIAGHDPLDSTSVEAPVPDVVA
Mvan_2115|M.vanbaalenii_PYR-1 YGLIACASSLDQGGPCARTVLDTALLHQVIAGHDPRDSTSVNAAVPDVVG
MAB_3341c|M.abscessus_ATCC_199 FGLVACASSLDQGGPCARTVLDTALLHQVIAGHDPRDSTSVDEPVPDVVA
MUL_0689|M.ulcerans_Agy99 SGVIGVASPQDMVGPMARTVTDVATLLTVMTGVDDSD-PTTRAGGAHTAT
MMAR_0937|M.marinum_M SGVIGVASPQDMVGPMARTVTDVATLLTVMTGVDDSD-PTTRAGGAHTAT
MSMEG_3400|M.smegmatis_MC2_155 TGFSPLVHFQDTPGPMARNVSDLAAVLDVIVGYDPTDSYTALATSGPEVG
*. . * ** .*.* * * : *:.* * * : .
MLBr_01702|M.leprae_Br4923 AAKAGES----GDLHGVRVGVVRQLRG-EGYQSGVLASFQAAVEQLIALG
MAV_3859|M.avium_104 AARAGAA----GDLKGVRVGVVKQLRG-EGYQPGVLASFEAAVEQLTALG
Mb3036c|M.bovis_AF2122/97 AARAGAV----GDLRGVRVGVVRQLHGGEGYQPGVLASFEAAVEQLTALG
Rv3011c|M.tuberculosis_H37Rv AARAGAV----GDLRGVRVGVVRQLHGGEGYQPGVLASFEAAVEQLTALG
TH_2639|M.thermoresistible__bu AARAGAS----GDLKGVRVGVVRQLRG-EGIQPGVLASFDAAVEQLAALG
Mflv_4246|M.gilvum_PYR-GCK AARTGAG----GDLTGVRIGVVKQLRSGEGYQAGVLASFNAAVDQLTALG
Mvan_2115|M.vanbaalenii_PYR-1 AARAGAR----GDLKGVRIGVVKQLRSGEGYQPGVLASFTAAVEQLTALG
MAB_3341c|M.abscessus_ATCC_199 AARAGAS----GDLKGVRVGVVTQLRG-DGYQPGVMASFDAAVEQLTALG
MUL_0689|M.ulcerans_Agy99 DYRRFLD---PAALQGARLGVARERFGA---HEATDALIEGALGQLAALG
MMAR_0937|M.marinum_M DYRRFLD---PAALQGARLGVARERFGA---HEATDALIEGALGQLAALG
MSMEG_3400|M.smegmatis_MC2_155 DYGEALDGVDADTLSGFRVGVLTDAFGAGDDQELTNSVVRAAIDRLRHHG
* * *:** : . : . : . .*: :* *
MLBr_01702|M.leprae_Br4923 ATVSE-VDCPHFDYALAAYYLILPSEVSSNLARFDAMRYGLRVGDDGSRS
MAV_3859|M.avium_104 AEVSE-VDCPHFEYALAAYYLILPSEVSSNLARFDAMRYGLRIGDDGSHS
Mb3036c|M.bovis_AF2122/97 AEVSE-VDCPHFDHALAAYYLILPSEVSSNLARFDAMRYGLRVGDDGTRS
Rv3011c|M.tuberculosis_H37Rv AEVSE-VDCPHFDHALAAYYLILPSEVSSNLARFDAMRYGLRVGDDGTRS
TH_2639|M.thermoresistible__bu AEVIE-VDCPNIDHALAAYYLIMPSEVSSNLARFDAMRYGLRVGDDGTRS
Mflv_4246|M.gilvum_PYR-GCK AEVTE-VDCPHFDYSLPAYYLILPSEVSSNLAKFDGMRYGLRAGDDGTHS
Mvan_2115|M.vanbaalenii_PYR-1 AEVSE-VDCPHFDHSLAAYYLILPSEVSSNLAKFDGMRYGLRVGDDGTTS
MAB_3341c|M.abscessus_ATCC_199 ADVVE-VSCPHFEYALPAYYLILPSEVSSNLARFDAMRYGMRVGDDGTHS
MUL_0689|M.ulcerans_Agy99 AEIVDPIQASSLPFFGDLELELFRYGLKASLNGYLGAHPRAAVGSLDEPI
MMAR_0937|M.marinum_M AEIVDPIQASSLPFFGDLELELFRYGLKASLNGYLGAHPRAAVGSLDELI
MSMEG_3400|M.smegmatis_MC2_155 TGVVDGIVLGNLETW-VAETSLYTIQSKFDLEKFLGSRRHTGPTTIREIV
: : : : : : . .* : . :
MLBr_01702|M.leprae_Br4923 AEEVIALTRAAGFGPEVKRRIMIGAYALSAGYYDSYYSQAQKVRTLIARD
MAV_3859|M.avium_104 AEEVMALTRAAGFGPEVKRRIMIGTYALSAGYYDAYYNQAQKVRTLIARD
Mb3036c|M.bovis_AF2122/97 AEEVMAMTRAAGFGPEVKRRIMIGTYALSAGYYDAYYNQAQKVRTLIARD
Rv3011c|M.tuberculosis_H37Rv AEEVMAMTRAAGFGPEVKRRIMIGTYALSAGYYDAYYNQAQKVRTLIARD
TH_2639|M.thermoresistible__bu AEEVMALTRAAGFGPEVKRRIMIGTYALSAGYYDAYYNQAQKVRTLVARD
Mflv_4246|M.gilvum_PYR-GCK AEEVMALTRAAGFGPEVKRRIMIGAYALSAGYYDAYYNQAQKVRTLIARD
Mvan_2115|M.vanbaalenii_PYR-1 AEEVMAMTRAAGFGAEVKRRIMIGTYALSAGYYDAYYNQAQKVRTLIARD
MAB_3341c|M.abscessus_ATCC_199 AEEVMALTRAAGFGPEVKRRIMIGTYALSAGYYDAYYGRAQKVRTLIKRD
MUL_0689|M.ulcerans_Agy99 AFNRAHAGQVMPYFGQEFLEQSQAKGDLTDSQYLRVRAELRRLAGADGID
MMAR_0937|M.marinum_M AFNRAHAGQVMPYFGQEFLEQSQAKGDLTDPQYLRVRAELRRLAGADGID
MSMEG_3400|M.smegmatis_MC2_155 DHGDFHP---LTDLLADIADGPSNPEDHPEYFKKRTRQEDWRRT----LL
. . :
MLBr_01702|M.leprae_Br4923 LDKAYRSVDVLVSPATPTTAFSLGEKADDPLAMYLFDLCTLPLNLAGHCG
MAV_3859|M.avium_104 LDAAYESVDVVVSPATPTTAFGLGEKVDDPLAMYLFDLCTLPLNLAGHCG
Mb3036c|M.bovis_AF2122/97 LDAAYRSVDVLVSPTTPTTAFRLGEKVDDPLAMYLFDLCTLPLNLAGHCG
Rv3011c|M.tuberculosis_H37Rv LDAAYRSVDVLVSPTTPTTAFRMGEKVDDPLAMYLFDLCTLPLNLAGHCG
TH_2639|M.thermoresistible__bu FDAAYEKVDVLVSPTTPTTAFPLGEKVDHPLAMYLFDLCTLPLNLAGHCG
Mflv_4246|M.gilvum_PYR-GCK LDAAYQKVDVLVSPATPTTAFRLGEKVDDPLSMYLFDLCTLPLNLAGHCG
Mvan_2115|M.vanbaalenii_PYR-1 LDEAYRSVDVLVSPATPSTAFRLGEKVDDPLAMYLFDLCTLPLNLAGHCG
MAB_3341c|M.abscessus_ATCC_199 LERAYEQVDVLVTPATPTTAFRLGEKVDDPLAMYQFDLCTLPLNLAGHCG
MUL_0689|M.ulcerans_Agy99 KALREHRLDAIVAPTEGSPAFAIDPVVGDNILPGGCSTPPAVAG---YPH
MMAR_0937|M.marinum_M KALREHRLDAIVAPTEGSPAFAIDPVVGDNILPGGCSTPPAVAG---YPH
MSMEG_3400|M.smegmatis_MC2_155 AKMADAEIDFLVYPTVQVPAPTRADLAAKRWTALNFPTNTVIASQTSLPA
:* :* *: .* . . . .
MLBr_01702|M.leprae_Br4923 MSVPSGLSSDDDLPVGLQIMAPALADDRLYRVGAAYEAARGPLPSAI---
MAV_3859|M.avium_104 MSVPSGLSPDDDLPVGLQIMAPALADDRLYRVGAAYEAARGPLRSAI---
Mb3036c|M.bovis_AF2122/97 MSVPSGLSPDDGLPVGLQIMAPALADDRLYRVGAAYEAARGPLLSAI---
Rv3011c|M.tuberculosis_H37Rv MSVPSGLSPDDGLPVGLQIMAPALADDRLYRVGAAYEAARGPLLSAI---
TH_2639|M.thermoresistible__bu MSVPSGLSPDDNLPVGLQIMAPALADDRLYRVGAAYEAARGPLPAAV---
Mflv_4246|M.gilvum_PYR-GCK MSVPSGLSADDNLPVGLQIMAPALADDRLYRVGAAYEAARGPLPTAL---
Mvan_2115|M.vanbaalenii_PYR-1 MSVPSGLSADDNLPVGLQIMAPALADDRLYRVGAAYEAARGPLPTAL---
MAB_3341c|M.abscessus_ATCC_199 MSVPSGLSADDGLPVGLQIMAPALADDRLYRVGAAYETARGALPAAL---
MUL_0689|M.ulcerans_Agy99 ICVPAGYFCG--LPVGLSLFAGAFQEPKLIGYAYAFEQATGVRRPPRFVP
MMAR_0937|M.marinum_M ICVPAGYFCG--LPVGLSLFAGAFQEPKLIGYAYAFEQATGVRRPPRFVP
MSMEG_3400|M.smegmatis_MC2_155 MSIPVGFTDSG-LPVGLEVVGRPLTERALLRFARAWELAEAPRRQPQIEA
:.:* * . *****.:.. .: : * . *:* * . .
MLBr_01702|M.leprae_Br4923 -------
MAV_3859|M.avium_104 -------
Mb3036c|M.bovis_AF2122/97 -------
Rv3011c|M.tuberculosis_H37Rv -------
TH_2639|M.thermoresistible__bu -------
Mflv_4246|M.gilvum_PYR-GCK -------
Mvan_2115|M.vanbaalenii_PYR-1 -------
MAB_3341c|M.abscessus_ATCC_199 -------
MUL_0689|M.ulcerans_Agy99 TLST---
MMAR_0937|M.marinum_M TLST---
MSMEG_3400|M.smegmatis_MC2_155 SKARVTM