For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PSQTPGVTELSPIADAYGLGLSEGELAEFVPHVSEVLRSWDLVDELYERERPDAVTRTWAEPEHNPFGAW YVTCDITESTDGPLAGRRVAVKDNTAVAGVPMMNGSAALEGFIPDTDATIISRMLAAGATIAGKAVCEDL CFSGASHTSATGPVRNPWDPTRTSGGSSSGSAALVAAGIVDIATGGDQGGSVRLPAAFTGIVGHKPTHGL VPYTGAFPIEQTLDHLGPMTRTVADAALLLGVLAGRDGLDPRQPTRVAVPDYVAELKRPVAGMRVGVLCE GFGHPEAVPGVDDAVRRAVDVLRAQGMQCENVSIPWHRHGRKIWDVIAVEGATTQMVDGNAYGMNWQGAY DPALIEFFGSRRQADGTLLPDTVKLVLLTGRHTIRRHHGAHYAKARNLVPRLRAAYDDALSRFDVLVMPT TPLVATPIPESDAPRDEYLARALEMMANTAGLDVSGHPACSVPAGLVDGLPTGMMIVGRHFDDATVLRVA HAYEEATGGFLPAKVSAP*
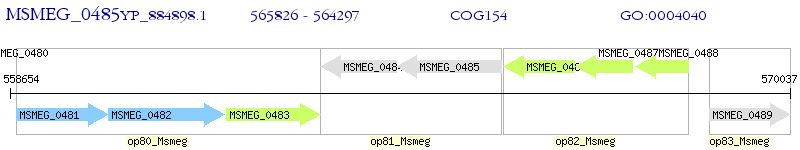
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0485 | - | - | 100% (509) | amidase |
| M. smegmatis MC2 155 | MSMEG_2365 | gatA | 5e-51 | 34.41% (433) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. smegmatis MC2 155 | MSMEG_1088 | gatA | 3e-50 | 34.91% (424) | glutamyl-tRNA(Gln)/aspartyl-tRNA(Asn) amidotransferase, A |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3036c | gatA | 3e-52 | 35.16% (438) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. gilvum PYR-GCK | Mflv_4246 | gatA | 2e-51 | 33.72% (430) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. tuberculosis H37Rv | Rv3011c | gatA | 4e-52 | 35.16% (438) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. leprae Br4923 | MLBr_01702 | gatA | 1e-48 | 33.57% (426) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. abscessus ATCC 19977 | MAB_3341c | gatA | 9e-52 | 35.31% (439) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. marinum M | MMAR_1702 | gatA | 3e-50 | 33.80% (432) | glutamyl-tRNA (Gln) amidotransferase (subunit A), GatA |
| M. avium 104 | MAV_3859 | gatA | 3e-48 | 33.72% (433) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. thermoresistible (build 8) | TH_2639 | - | 1e-51 | 35.39% (438) | PUTATIVE glutamyl-tRNA (Gln) amidotransferase (subunit A), GatA |
| M. ulcerans Agy99 | MUL_1939 | gatA | 2e-50 | 33.80% (432) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. vanbaalenii PYR-1 | Mvan_2115 | gatA | 4e-51 | 34.64% (433) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3036c|M.bovis_AF2122/97 ------MTDIIRSDAATLAAKIAIKEVSSTEITRACLDQIEATDETYHAF
Rv3011c|M.tuberculosis_H37Rv ------MTDIIRSDAATLAAKIAIKEVSSAEITRACLDQIEATDETYHAF
MMAR_1702|M.marinum_M ------MTDLIRSDAATLAAKIAAKEVSATELTQACLDQIEATDDRYHAF
MUL_1939|M.ulcerans_Agy99 ------MTDLIRSDAATLAAKIAAKEVSATELTQACLDQIEATDDRYHAF
MLBr_01702|M.leprae_Br4923 --MIGSLHEIIRLDASTLAAKIVAKELSSVEITQACLDQIEATDDTYRAF
MAV_3859|M.avium_104 ------MNEIIRSDAATLAARIAAKELSSVEATQACLDQIAATDERYHAF
Mflv_4246|M.gilvum_PYR-GCK ------MSELIKLDAATLADKISSKEVSSAEVTQAFLDQIAATDGDYHAF
Mvan_2115|M.vanbaalenii_PYR-1 ------MTDLIKLDAATLAAKIAAREVSATEVTQACLDQIAATDAEYHAF
TH_2639|M.thermoresistible__bu MSQGKSTDDLIRLDAATLGAKIAAKEISSTELTQACLDRIAATDDRYNAF
MAB_3341c|M.abscessus_ATCC_199 ------MTDLIRQDAAALAALIHSGEVSSVEVTRAHLDQIASTDETYRAF
MSMEG_0485|M.smegmatis_MC2_155 --MPSQTPGVTELSPIADAYGLGLSEGELAEFVPHVSEVLRSWDLVDELY
: . .. : . : * . .* . : : : * . :
Mb3036c|M.bovis_AF2122/97 LHVAADEALAAAAAVDK--------QVAAGEPLPSALAGVPLALKDVFTT
Rv3011c|M.tuberculosis_H37Rv LHVAADEALAAAAAIDK--------QVAAGEPLPSALAGVPLALKDVFTT
MMAR_1702|M.marinum_M LHIGAHEALSAAAAVDT--------ALAAGERLPSALAGVPLALKDVFTT
MUL_1939|M.ulcerans_Agy99 LHIGAHEALSAAAAVDT--------ALAAGERLPSALAGVPLALKDVFTT
MLBr_01702|M.leprae_Br4923 LHVAAEKALSAAAAVDK--------AVAAGGQLSSTLAGVPLALKDVFTT
MAV_3859|M.avium_104 LHVAADEALGAAAVVDE--------AVAAGERLPSPLAGVPLALKDVFTT
Mflv_4246|M.gilvum_PYR-GCK LHVGAEQALAAAQTVDR--------AVAAGEHLPSPLAGVPLALKDVFTT
Mvan_2115|M.vanbaalenii_PYR-1 LHVAGDQALAAAATVDKAIH----EATAAGERLPSPLAGVPLALKDVFTT
TH_2639|M.thermoresistible__bu LHVAANKALSAAARVDA--------AVAAGEELPSPLAGVPLALKDVFTT
MAB_3341c|M.abscessus_ATCC_199 LHVAGEQALAAAKDVDD--------AIAAGS-VPSGLAGVPLALKDVFTT
MSMEG_0485|M.smegmatis_MC2_155 ERERPDAVTRTWAEPEHNPFGAWYVTCDITESTDGPLAGRRVAVKDNTAV
: . . : : . *** :*:** :.
Mb3036c|M.bovis_AF2122/97 SDMPTTCGSKILEGWRSPYDATLTARLRAAGIPILGKTNMDEFAMGSSTE
Rv3011c|M.tuberculosis_H37Rv SDMPTTCGSKILEGWRSPYDATLTARLRAAGIPILGKTNMDEFAMGSSTE
MMAR_1702|M.marinum_M VDMPTTCGSKILEGWRSPYDATLTLRLRAAGIPILGKTNMDEFAMGSSTE
MUL_1939|M.ulcerans_Agy99 VDMPTTCGSKILEGWRSPYDATLTLRLRAAGIPILGKTNMDEFAMGSSTE
MLBr_01702|M.leprae_Br4923 VDMPTTCGSKILQGWHSPYDATVTTRLRAAGIPILGKTNMDEFAMGSSTE
MAV_3859|M.avium_104 VDMPTTCGSKILQGWRSPYDATVTTKLRAAGIPILGKTNMDEFAMGSSTE
Mflv_4246|M.gilvum_PYR-GCK TDMPTTCGSKVLEGWTSPYDATVTAKLRSAGIPILGKTNMDEFAMGSSTE
Mvan_2115|M.vanbaalenii_PYR-1 TDMPTTCGSKILEGWTSPYDATVTAKLRAAGIPILGKTNMDEFAMGSSTE
TH_2639|M.thermoresistible__bu IDMPTTCGSKILDGWQAPYDATVTVALRAAGIPILGKTNLDEFAMGSSTE
MAB_3341c|M.abscessus_ATCC_199 RDMPTTCGSKILENWVPPYDATVTARLRGAGIPILGKTNMDEFAMGSSTE
MSMEG_0485|M.smegmatis_MC2_155 AGVPMMNGSAALEGFIPDTDATIISRMLAAGATIAGKAVCEDLCFSGASH
.:* ** *:.: . ***: : .** .* **: :::.:..::.
Mb3036c|M.bovis_AF2122/97 NSAYGPTRNPWNLDRVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAA
Rv3011c|M.tuberculosis_H37Rv NSAYGPTRNPWNLDRVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAA
MMAR_1702|M.marinum_M NSAYGPTRNPWNLDRVPGGSGGGSAAALAAYQAPLAIGSDTGGSIRQPAA
MUL_1939|M.ulcerans_Agy99 NSAYGPTRNPWNLDRVPGGSGGGSAAALAAYQAPLAIGSDTGGSIRQPAA
MLBr_01702|M.leprae_Br4923 NSAYGPTRNPWNVDRVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAA
MAV_3859|M.avium_104 NSAYGPTRNPWNVERVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAA
Mflv_4246|M.gilvum_PYR-GCK NSAYGPTRNPWDTERVPGGSGGGSAAALAAFQAPLAIGTDTGGSIRQPAA
Mvan_2115|M.vanbaalenii_PYR-1 NSAYGPTRNPWNVDCVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAA
TH_2639|M.thermoresistible__bu NSAFGPTRNPWDVNRVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAA
MAB_3341c|M.abscessus_ATCC_199 NSAYGPTRNPWDVERVPGGSGGGSAAALAAFQAPLAIGTDTGGSIRQPAA
MSMEG_0485|M.smegmatis_MC2_155 TSATGPVRNPWDPTRTSGGSSSGSAALVAAGIVDIATGGDQGGSVRLPAA
.** **.****: ..***..**** :** . :* * * ***:* ***
Mb3036c|M.bovis_AF2122/97 LTATVGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTALLHQVIAGHD
Rv3011c|M.tuberculosis_H37Rv LTATVGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTALLHQVIAGHD
MMAR_1702|M.marinum_M LTATVGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTAMLHQVIAGHD
MUL_1939|M.ulcerans_Agy99 LTATVGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTAMLHQVIAGHD
MLBr_01702|M.leprae_Br4923 LTATVGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTALLHAVIAGHD
MAV_3859|M.avium_104 LTATVGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTALLHQVIAGHD
Mflv_4246|M.gilvum_PYR-GCK LTATVGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTALLHQVIAGHD
Mvan_2115|M.vanbaalenii_PYR-1 LTATVGVKPTYGTVSRYGLIACASSLDQGGPCARTVLDTALLHQVIAGHD
TH_2639|M.thermoresistible__bu LTATVGVKPTYGTVSRYGLVACASSLDQGGPCGRTVLDTALLHQVIAGHD
MAB_3341c|M.abscessus_ATCC_199 LTATVGVKPTYGTVSRFGLVACASSLDQGGPCARTVLDTALLHQVIAGHD
MSMEG_0485|M.smegmatis_MC2_155 FTGIVGHKPTHGLVPYTGAFPIEQTLDHLGPMTRTVADAALLLGVLAGRD
:*. ** ***:* *. * .. .:**: ** *** *:*:* *:**:*
Mb3036c|M.bovis_AF2122/97 PRDS-TSVDAEVPDVVGAARAGAVGDLRGVRVGVVRQLHGGEGYQPGVLA
Rv3011c|M.tuberculosis_H37Rv PRDS-TSVDAEVPDVVGAARAGAVGDLRGVRVGVVRQLHGGEGYQPGVLA
MMAR_1702|M.marinum_M AKDS-TSLETEIPDVVGAAKAGASGDLRGVRIGVVKQLRG-DGYQPGVLA
MUL_1939|M.ulcerans_Agy99 AKDS-TSLETEIPDVVGAAKAGASGDLRGVRIGVVKQLRG-DGYQPGVLA
MLBr_01702|M.leprae_Br4923 ARDS-TSVEAAVPDIVGAAKAGESGDLHGVRVGVVRQLRG-EGYQSGVLA
MAV_3859|M.avium_104 IRDS-TSVDAPVPDVVGAARAGAAGDLKGVRVGVVKQLRG-EGYQPGVLA
Mflv_4246|M.gilvum_PYR-GCK PLDS-TSVEAPVPDVVAAARTGAGGDLTGVRIGVVKQLRSGEGYQAGVLA
Mvan_2115|M.vanbaalenii_PYR-1 PRDS-TSVNAAVPDVVGAARAGARGDLKGVRIGVVKQLRSGEGYQPGVLA
TH_2639|M.thermoresistible__bu AKDS-TSLTAPVPDVVAAARAGASGDLKGVRVGVVRQLRG-EGIQPGVLA
MAB_3341c|M.abscessus_ATCC_199 PRDS-TSVDEPVPDVVAAARAGASGDLKGVRVGVVTQLRG-DGYQPGVMA
MSMEG_0485|M.smegmatis_MC2_155 GLDPRQPTRVAVPDYVAELKRPVAG----MRVGVLCEGFGHPEAVPGVDD
*. . :** *. : * :*:**: : . .**
Mb3036c|M.bovis_AF2122/97 SFEAAVEQLTALGAEVSEVDCPHFDHALAAYYLILPSEVSSNLARFDAMR
Rv3011c|M.tuberculosis_H37Rv SFEAAVEQLTALGAEVSEVDCPHFDHALAAYYLILPSEVSSNLARFDAMR
MMAR_1702|M.marinum_M SFEAAVAQLTALGAEVSEVDCPHFDHALAAYYLILPSEVSSNLARFDAMR
MUL_1939|M.ulcerans_Agy99 SFEAAVAQLTALGAEVSEVDCPHFDHALAAYYLILPSEVSSNLARFDAMR
MLBr_01702|M.leprae_Br4923 SFQAAVEQLIALGATVSEVDCPHFDYALAAYYLILPSEVSSNLARFDAMR
MAV_3859|M.avium_104 SFEAAVEQLTALGAEVSEVDCPHFEYALAAYYLILPSEVSSNLARFDAMR
Mflv_4246|M.gilvum_PYR-GCK SFNAAVDQLTALGAEVTEVDCPHFDYSLPAYYLILPSEVSSNLAKFDGMR
Mvan_2115|M.vanbaalenii_PYR-1 SFTAAVEQLTALGAEVSEVDCPHFDHSLAAYYLILPSEVSSNLAKFDGMR
TH_2639|M.thermoresistible__bu SFDAAVEQLAALGAEVIEVDCPNIDHALAAYYLIMPSEVSSNLARFDAMR
MAB_3341c|M.abscessus_ATCC_199 SFDAAVEQLTALGADVVEVSCPHFEYALPAYYLILPSEVSSNLARFDAMR
MSMEG_0485|M.smegmatis_MC2_155 AVRRAVDVLRAQGMQCENVSIPWHRHGRKIWDVIAVEGATT--QMVDGNA
:. ** * * * :*. * :. : :* . .:: .*.
Mb3036c|M.bovis_AF2122/97 YGLRVGDDGTRSAEEVMAMTRAAG--FGPEVKRRIMIGTYALSAGYYDAY
Rv3011c|M.tuberculosis_H37Rv YGLRVGDDGTRSAEEVMAMTRAAG--FGPEVKRRIMIGTYALSAGYYDAY
MMAR_1702|M.marinum_M YGLRIGDDGTHSAEEVMAMTRAAG--FGPEVKRRIMIGAYALSAGYYDAY
MUL_1939|M.ulcerans_Agy99 YGLRIGDDGTHSAEEVMAMTRAAG--FGPEVKRRIMIGAYALSAGYYDAY
MLBr_01702|M.leprae_Br4923 YGLRVGDDGSRSAEEVIALTRAAG--FGPEVKRRIMIGAYALSAGYYDSY
MAV_3859|M.avium_104 YGLRIGDDGSHSAEEVMALTRAAG--FGPEVKRRIMIGTYALSAGYYDAY
Mflv_4246|M.gilvum_PYR-GCK YGLRAGDDGTHSAEEVMALTRAAG--FGPEVKRRIMIGAYALSAGYYDAY
Mvan_2115|M.vanbaalenii_PYR-1 YGLRVGDDGTTSAEEVMAMTRAAG--FGAEVKRRIMIGTYALSAGYYDAY
TH_2639|M.thermoresistible__bu YGLRVGDDGTRSAEEVMALTRAAG--FGPEVKRRIMIGTYALSAGYYDAY
MAB_3341c|M.abscessus_ATCC_199 YGMRVGDDGTHSAEEVMALTRAAG--FGPEVKRRIMIGTYALSAGYYDAY
MSMEG_0485|M.smegmatis_MC2_155 YGMNWQGAYDPALIEFFGSRRQADGTLLPDTVKLVLLTGRHTIRRHHGAH
**:. . : *.:. * *. : .:. : ::: ::.::
Mb3036c|M.bovis_AF2122/97 YNQAQKVRTLIARDLDAAYRSVDVLVSPTTPTTAFRLGEK---VDDPLAM
Rv3011c|M.tuberculosis_H37Rv YNQAQKVRTLIARDLDAAYRSVDVLVSPTTPTTAFRMGEK---VDDPLAM
MMAR_1702|M.marinum_M YNQAQKVRTLIARDLDEAYQSVDVLVSPATPTTAFPLGEK---VDDPLAM
MUL_1939|M.ulcerans_Agy99 YNQAQKIRTLIARDLDEAYQSVDVLVSPATPTTAFPLGEK---VDDPLAM
MLBr_01702|M.leprae_Br4923 YSQAQKVRTLIARDLDKAYRSVDVLVSPATPTTAFSLGEK---ADDPLAM
MAV_3859|M.avium_104 YNQAQKVRTLIARDLDAAYESVDVVVSPATPTTAFGLGEK---VDDPLAM
Mflv_4246|M.gilvum_PYR-GCK YNQAQKVRTLIARDLDAAYQKVDVLVSPATPTTAFRLGEK---VDDPLSM
Mvan_2115|M.vanbaalenii_PYR-1 YNQAQKVRTLIARDLDEAYRSVDVLVSPATPSTAFRLGEK---VDDPLAM
TH_2639|M.thermoresistible__bu YNQAQKVRTLVARDFDAAYEKVDVLVSPTTPTTAFPLGEK---VDHPLAM
MAB_3341c|M.abscessus_ATCC_199 YGRAQKVRTLIKRDLERAYEQVDVLVTPATPTTAFRLGEK---VDDPLAM
MSMEG_0485|M.smegmatis_MC2_155 YAKARNLVPRLRAAYDDALSRFDVLVMPTTPLVATPIPESDAPRDEYLAR
* :*::: . : : * .**:* *:** .* : *. *. *:
Mb3036c|M.bovis_AF2122/97 YLFDLCTLP-LNLAGHCGMSVPSGLSPDDGLPVGLQIMAPALADDRLYRV
Rv3011c|M.tuberculosis_H37Rv YLFDLCTLP-LNLAGHCGMSVPSGLSPDDGLPVGLQIMAPALADDRLYRV
MMAR_1702|M.marinum_M YLFDLCTLP-LNLAGHCGMSVPSGLSPDDGLPVGLQIMAPALADDRLYRV
MUL_1939|M.ulcerans_Agy99 YLFDLCTLP-LNLAGHCGMSVPSGLSPDDGLPVGLQIMAPALADDRLYRV
MLBr_01702|M.leprae_Br4923 YLFDLCTLP-LNLAGHCGMSVPSGLSSDDDLPVGLQIMAPALADDRLYRV
MAV_3859|M.avium_104 YLFDLCTLP-LNLAGHCGMSVPSGLSPDDDLPVGLQIMAPALADDRLYRV
Mflv_4246|M.gilvum_PYR-GCK YLFDLCTLP-LNLAGHCGMSVPSGLSADDNLPVGLQIMAPALADDRLYRV
Mvan_2115|M.vanbaalenii_PYR-1 YLFDLCTLP-LNLAGHCGMSVPSGLSADDNLPVGLQIMAPALADDRLYRV
TH_2639|M.thermoresistible__bu YLFDLCTLP-LNLAGHCGMSVPSGLSPDDNLPVGLQIMAPALADDRLYRV
MAB_3341c|M.abscessus_ATCC_199 YQFDLCTLP-LNLAGHCGMSVPSGLSADDGLPVGLQIMAPALADDRLYRV
MSMEG_0485|M.smegmatis_MC2_155 ALEMMANTAGLDVSGHPACSVPAGLVDG--LPTGMMIVGRHFDDATVLRV
:.. . *:::** . ***:** . **.*: *:. : * : **
Mb3036c|M.bovis_AF2122/97 GAAYEAARGPLLSAI----
Rv3011c|M.tuberculosis_H37Rv GAAYEAARGPLLSAI----
MMAR_1702|M.marinum_M GAAYEAARGPLPSAI----
MUL_1939|M.ulcerans_Agy99 GAAYEAARGPLPSAI----
MLBr_01702|M.leprae_Br4923 GAAYEAARGPLPSAI----
MAV_3859|M.avium_104 GAAYEAARGPLRSAI----
Mflv_4246|M.gilvum_PYR-GCK GAAYEAARGPLPTAL----
Mvan_2115|M.vanbaalenii_PYR-1 GAAYEAARGPLPTAL----
TH_2639|M.thermoresistible__bu GAAYEAARGPLPAAV----
MAB_3341c|M.abscessus_ATCC_199 GAAYETARGALPAAL----
MSMEG_0485|M.smegmatis_MC2_155 AHAYEEATGGFLPAKVSAP
. *** * * : .*