For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NRVTDEAVELSGSDARRIAFAAFVGTALEWYDYFLFGTAAALVFNRLFFTDLDPAAATLASFATFGVGFA ARPIGAVIFGYIGDRYGRRPALLATIIMIGCATGLVGVLPDYAAVGIAAPAMLAILRLVQGLAVGGEWGG ATTMAIEHSPIKQRGRFAALVQIGSPVGTLVSSGAFALVLMLPSESFDSWGWRLPFLAAFPLLGVALWIR LTMEESPVFKRLAELEAAAEVPVLQLVRNAGGRLLVAVAAALLGVGGFYIMTTFVVSYGSNVLAVDRQVM VNATLVAAVLQIAVTIFAGRLSERFGPGRMTVAGAVTTAIAAFPLFWLIDTRNAWAITAAVACGIALITL AYAVTGALLAELFPPALRYSGVSLGYNLAGALSGFLPLIATALLAVGDGASWPAALVLIGICAITAIGGL AGERMRLRHDPVEHQPATS*
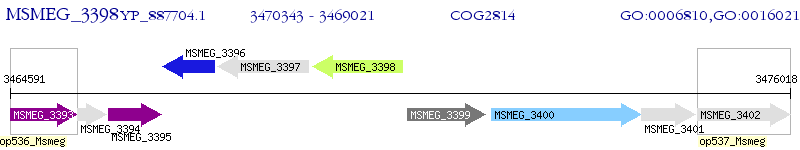
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3398 | - | - | 100% (440) | integral membrane transport protein |
| M. smegmatis MC2 155 | MSMEG_4122 | - | 4e-93 | 42.00% (419) | major facilitator family protein transporter |
| M. smegmatis MC2 155 | MSMEG_0716 | - | 5e-92 | 41.34% (404) | major facilitator superfamily protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1232 | - | 1e-71 | 36.10% (410) | integral membrane transport protein |
| M. gilvum PYR-GCK | Mflv_2383 | - | 2e-88 | 44.09% (406) | major facilitator transporter |
| M. tuberculosis H37Rv | Rv1200 | - | 1e-71 | 36.10% (410) | integral membrane transport protein |
| M. leprae Br4923 | MLBr_01222 | - | 0.0001 | 25.40% (126) | hypothetical protein MLBr_01222 |
| M. abscessus ATCC 19977 | MAB_2481 | - | 1e-83 | 41.49% (388) | putative integral membrane transporter |
| M. marinum M | MMAR_4238 | - | 2e-66 | 36.71% (395) | integral membrane transport protein |
| M. avium 104 | MAV_4627 | - | 9e-83 | 39.00% (418) | major facilitator superfamily protein |
| M. thermoresistible (build 8) | TH_3714 | - | 7e-92 | 43.51% (439) | PUTATIVE major facilitator superfamily MFS_1 |
| M. ulcerans Agy99 | MUL_0949 | - | 4e-66 | 36.46% (395) | integral membrane transport protein |
| M. vanbaalenii PYR-1 | Mvan_2899 | - | 2e-85 | 40.55% (397) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1232|M.bovis_AF2122/97 -----------------------------MKRVALACLVGSAIEFYDFLI
Rv1200|M.tuberculosis_H37Rv -----------------------------MKRVALACLVGSAIEFYDFLI
MMAR_4238|M.marinum_M -----------------------------MSRVALACVVGSTVEYYDFCI
MUL_0949|M.ulcerans_Agy99 -----------------------------MSRVALACVVGSTVEYYDFCI
Mvan_2899|M.vanbaalenii_PYR-1 ------------------MNEASVADSALLRRVALSSLLGTAIEYYDFLV
Mflv_2383|M.gilvum_PYR-GCK MGP----------------GGRT---RVSPRRVAIASFVGTTVEFYDFLI
TH_3714|M.thermoresistible__bu MSA----------------EAATPTSRVTPRRVAVASFIGTTVEFYDFLI
MAV_4627|M.avium_104 MAATLPAPAPAGLADGHPLDESAEQQRRRLRIVVAASLLGTTVEWYDFFL
MAB_2481|M.abscessus_ATCC_1997 -------MTTTSAVPAVHAGGPADGDAPAARTVALSCMVGTTIEWYDFYL
MSMEG_3398|M.smegmatis_MC2_155 --------------MNRVTDEAVELSGSDARRIAFAAFVGTALEWYDYFL
MLBr_01222|M.leprae_Br4923 --------------------------------------------------
Mb1232|M.bovis_AF2122/97 YGTAAALVFPTVFFPHLDPTVAAVASMGTFAVAFLSRPFGAAVFGYFGDR
Rv1200|M.tuberculosis_H37Rv YGTAAALVFPTVFFPHLDPTVAAVASMGTFAVAFLSRPFGAAVFGYFGDR
MMAR_4238|M.marinum_M YGTAAALVFPAVFYPGLSPALATIASMGTFATAFLSRPLGAAVFGHFGDR
MUL_0949|M.ulcerans_Agy99 YGTAAALVFPAVFYPGLSPALATIASMGTFATAFLSRPLGAAVFGHFGDR
Mvan_2899|M.vanbaalenii_PYR-1 YGTMSALVFGLVFFPDSDPAVATIAAFGTLAAGYVARPVGGIVFGHFGDR
Mflv_2383|M.gilvum_PYR-GCK YGTAAALVFPKLFFPNASPAAGILLSFATFGVGFIARPLGGIVFGHFGDR
TH_3714|M.thermoresistible__bu YGTAAALVFPKLFFPTASPASGVLLSFATFGVGFFARPLGGIVFGHFGDR
MAV_4627|M.avium_104 YATAAGLVFNKVFFPNESSLVGTLLAFATFAVGFVMRPIGGLVFGHIGDR
MAB_2481|M.abscessus_ATCC_1997 YATAAALVLKPLFFPSVSPTAGTLASFATYAAGFGARPVGAVIAGHLGDR
MSMEG_3398|M.smegmatis_MC2_155 FGTAAALVFNRLFFTDLDPAAATLASFATFGVGFAARPIGAVIFGYIGDR
MLBr_01222|M.leprae_Br4923 ----------------------MTEHCASDISDVSCPPRGRVIVG-----
.: * * : *
Mb1232|M.bovis_AF2122/97 LGRKKTLVATLLIMGLATVTVGLVPTTVAIGAAAPLILTTMRLLQGFAVG
Rv1200|M.tuberculosis_H37Rv LGRKKTLVATLLIMGLATVTVGLVPTTVAIGAAAPLILTTMRLLQGFAVG
MMAR_4238|M.marinum_M LGRKKTLVATLLIMALSTVTVGVVPSTATIGVSAPLILVALRLLQGFAVG
MUL_0949|M.ulcerans_Agy99 LGRKKTLVATLLIMALSTVTVGVVPSTATIGVSAPLILVALRLLQGFAVG
Mvan_2899|M.vanbaalenii_PYR-1 IGRKSMLVLTMTLMGTASFLIGLLPSFAAVGVAAPILLVALRVIQGVAIG
Mflv_2383|M.gilvum_PYR-GCK VGRRRMLVYSLVGMGVSTVLIGVLPTYAQIGLAAPILLTVLRLAQGFAVG
TH_3714|M.thermoresistible__bu LGRKRMLVYSLVGMGVATVLIGLLPTYAQIGMAAPALLTLLRLAQGFAMG
MAV_4627|M.avium_104 IGRKRSLALTMLIMGGATALIGVLPTAAQIGAWAPVLLLVLRVLQGFALG
MAB_2481|M.abscessus_ATCC_1997 VGRKAVLVGALLLMGVATAVIGALPTYSEAGVLAPAALATLRVLQGLAAG
MSMEG_3398|M.smegmatis_MC2_155 YGRRPALLATIIMIGCATGLVGVLPDYAAVGIAAPAMLAILRLVQGLAVG
MLBr_01222|M.leprae_Br4923 ------------------VVLGLAGTGALIGGLWAWIAPPIHAVVGLTRT
:* * . :: *.:
Mb1232|M.bovis_AF2122/97 GEWAGSALLSAEYAPASKRGWYGMFTVVGGGIALVLTSLTFLGVNYTIGE
Rv1200|M.tuberculosis_H37Rv GEWAGSALLSAEYAPASKRGWYGMFTVVGGGIALVLTSLTFLGVNYTIGE
MMAR_4238|M.marinum_M GEWAGSALLSIESAPAHKRGYYGMFTVLGGGVALVVSGLTFLGVNYTIGE
MUL_0949|M.ulcerans_Agy99 GEWAGSALLSIESAPAHKRGYYGMFTVLGGGVALVVSGLTFLGVNYTIGE
Mvan_2899|M.vanbaalenii_PYR-1 GEWGGATLMVTEHAEAHRRGFWNGVMQMGSPIGSLLSVAVVTVITLLPDE
Mflv_2383|M.gilvum_PYR-GCK GEWGGATLMAVEHATADRRGFYGAFPQMGAPAQTAAAT-LAFYLAARLPD
TH_3714|M.thermoresistible__bu GEWGGATLMAVEHASADRKGFYGAFPQMGAPAGTATAT-LAFFAVSRLPD
MAV_4627|M.avium_104 GEWGGAVLLAVEHSPGDRRGRYGAVPQVGLALGLALGTGIFAFLQIVLGP
MAB_2481|M.abscessus_ATCC_1997 AEWGGAALLAVEHAPVGRRGLFGSFTQLGSPAGMLLATGVFGLTRALTGP
MSMEG_3398|M.smegmatis_MC2_155 GEWGGATTMAIEHSPIKQRGRFAALVQIGSPVGTLVSSGAFALVLMLPSE
MLBr_01222|M.leprae_Br4923 GERG---------------------------------------HDYLGNE
.* .
Mb1232|M.bovis_AF2122/97 SSPTFMQWGWRIPFLVSAALIAVALYVRFNIDETPVFAR--ERADEKTRL
Rv1200|M.tuberculosis_H37Rv SSPTFMQWGWRIPFLVSAALIAVALYVRFNIDETPVFAR--ERADEKTRL
MMAR_4238|M.marinum_M SSSAFLQWGWRVPFLLSALLIAFALYVRLEINETPVFARAMAQADDQEST
MUL_0949|M.ulcerans_Agy99 SSSALLQWGWRVPFLLSALLIAFALYVRLEINETPVFARAMAQADDQEST
Mvan_2899|M.vanbaalenii_PYR-1 S---FLSWGWRIPFLVSLLLLAVGIYVRLSVTESPVFEA----ARKEAAG
Mflv_2383|M.gilvum_PYR-GCK DQ--FLAWGWRIPFLVSAILIVIGLVVRLTVTESPHFAE------LQRRS
TH_3714|M.thermoresistible__bu GQ--FLSWGWRLPFLLSAVLIGIGLAVRLTLTESPDFAA------VQARS
MAV_4627|M.avium_104 AR--FLSYGWRIGFLLSVLLVAIGIVVRLRVEETPAFRE------LQDLQ
MAB_2481|M.abscessus_ATCC_1997 EA--FLAFGWRIPFLSSIVLVVVGLAIRLRLRDSVVFRK------VLERG
MSMEG_3398|M.smegmatis_MC2_155 S---FDSWGWRLPFLAAFPLLGVALWIRLTMEESPVFKR------LAELE
MLBr_01222|M.leprae_Br4923 SEHFFVAPCLMLGLLTVLAVTASVLAWQLRQHRGP---------------
: : :* : : ::
Mb1232|M.bovis_AF2122/97 GPAETPIAQVLR--RQRREIVLAAGSAVCCFGFVYLASTYLASYAQTRLG
Rv1200|M.tuberculosis_H37Rv GPAETPIAQVLR--RQRREIVLAAGSAVCCFGFVYLASTYLASYAQTRLG
MMAR_4238|M.marinum_M AAARAPLAAVLC--RQRREIVLAAGAMLAAFGCLYLASTFLPSYAQLHLG
MUL_0949|M.ulcerans_Agy99 AAARAPLAAVLC--RQRREIVLAAGAMLAAFGCLYLASTFLPSYAQLHLG
Mvan_2899|M.vanbaalenii_PYR-1 NAPRVPFVEILR--RPR-TLVLACAVGIGPFALTALISTYMISYATA-IG
Mflv_2383|M.gilvum_PYR-GCK EVTRTPIVAAFR--RHWRQILLVAGAYLSQGIFAYICVAYLVSYATTVAE
TH_3714|M.thermoresistible__bu AVHRMPIAEAFR--RHRRQILLVAGAYLSQGVFAYICVAYLVSYGTSVAG
MAV_4627|M.avium_104 AASTVPIRDILRERRSRRNTVLGLLSRWAEGSAFNTWGVFAISYATGALG
MAB_2481|M.abscessus_ATCC_1997 EAARLPVVEVLR--TQSRNVLLTTGLRLSQIALFVLLTTFSLTYLQGSFG
MSMEG_3398|M.smegmatis_MC2_155 AAAEVPVLQLVR--NAGGRLLVAVAAALLGVGGFYIMTTFVVSYGSNVLA
MLBr_01222|M.leprae_Br4923 ---------------------------------------------GMVIG
Mb1232|M.bovis_AF2122/97 YSRGSILFDSVLGGLLCIVFTALSSALCDQLGRRRVLLAGWAVALPWSLL
Rv1200|M.tuberculosis_H37Rv YSRGSILFDSVLGGLLCIVFTALSSALCDQLGRRRVLLAGWAVALPWSLL
MMAR_4238|M.marinum_M YSRNLVLLIGVLGGLVCIGFVALSAILCDRFGRRRVMLVGWAVGLFWSPL
MUL_0949|M.ulcerans_Agy99 YSRNLVLLIGVLGGLVCIGFVALSAILCDRFGRRRVMLVGWAVGLFWSPL
Mvan_2899|M.vanbaalenii_PYR-1 YHRTDVMTALIFTSLTGLIAIPFFSALSDHVGRRLVMVAGAIGIIGYAWP
Mflv_2383|M.gilvum_PYR-GCK IPRDWALLGVFLGAVVAVATYPLFGALSDRVGRKPLYLAGVIAMALAVVP
TH_3714|M.thermoresistible__bu IDRTATLFGVSVAAVLAVVMYPLFGSLSDTFGRKPLFLAGVIAMGVSIGP
MAV_4627|M.avium_104 LNRVSVLIAVTIAALLMAVLLPVSGVLTDRFGARRVYLAGIACYGLAAFP
MAB_2481|M.abscessus_ATCC_1997 GDSQIGLTAVLISSALGILSTPVWSALSDRVGRRPLYLFGSAAGPLALGL
MSMEG_3398|M.smegmatis_MC2_155 VDRQVMVNATLVAAVLQIAVTIFAGRLSERFGPGRMTVAGAVTTAIAAFP
MLBr_01222|M.leprae_Br4923 LAIGLMICAATAAAVGALLVWMRYGALN--FDAVPLSYDHKVAHVIQAPP
: . . * .. :
Mb1232|M.bovis_AF2122/97 VMPLIDSGSPSLFAVAVVGMY-AIGGFGFGPTASFIPELFATSYRYTGSA
Rv1200|M.tuberculosis_H37Rv VMPLIDSGSPSLFAVAVVGMY-AIGGFGFGPTASFIPELFATSYRYTGSA
MMAR_4238|M.marinum_M VMPLIGSGDTVGFTVALLGLY-AVAASGFGPAASLIPELFATRYRYTGTA
MUL_0949|M.ulcerans_Agy99 VMPLIGSGDTVGFTVALLGLY-AVAASGFGPAASLIPELFATRYRYTGTA
Mvan_2899|M.vanbaalenii_PYR-1 FYALVDMRSQAYLTAAMVIAQ-LIQSMMYAPLGALFSEMFGTTVRYTGAS
Mflv_2383|M.gilvum_PYR-GCK TFALINTGRPVMFVLALVLIFGLAMAPAAGVTGALFSMAFDADVRYSGVS
TH_3714|M.thermoresistible__bu AFALIDTGRPALFLVALVLVFGLAMAPAAGVTGALFAMTFDADVRYSGVS
MAV_4627|M.avium_104 AFALFGTKKLLWFGLAMVIVFGVVHALFYGAQGTLYSALYPTNTRYTGLS
MAB_2481|M.abscessus_ATCC_1997 FFLAAGSGSKVLVVLSIVFGINVVHDAMYGPQAAWFAELFDTRVRYSGAS
MSMEG_3398|M.smegmatis_MC2_155 LFWLIDTRN-AWAITAAVACGIALITLAYAVTGALLAELFPPALRYSGVS
MLBr_01222|M.leprae_Br4923 VFFAHGLLQVAATVLWPAGIAALVYAVLAAANG-----------------
. . .
Mb1232|M.bovis_AF2122/97 LAANLAGVAGGALPPVIAGALVATYG-SWAIGVMLAI-LALISLVCTYRL
Rv1200|M.tuberculosis_H37Rv LAANLAGVAGGALPPVIAGALVATYG-SWAIGVMLAI-LALISLVCTYRL
MMAR_4238|M.marinum_M LALNVAGIVGGAAPPLIAGTLLASYG-SSAVGLMMTA-LVAASLLCTYLL
MUL_0949|M.ulcerans_Agy99 LALNVAGIVGGAAPPLIAGTLLASYG-SSAVGLMMTA-LVAASLLCTYLL
Mvan_2899|M.vanbaalenii_PYR-1 MGYQLAALLGAGFTPLIASALHAQEV-SRTPLVALAAGCGLVTVVAVWRI
Mflv_2383|M.gilvum_PYR-GCK VGYTLSQVLGSAFAPTIATALYAATGSSNSIAA-YLIAASAVSAVAVCFM
TH_3714|M.thermoresistible__bu IGYTLSQVLGSAFAPTIAAALYAATESSGSIVA-YLIGVSVISAVSVCLL
MAV_4627|M.avium_104 FVYQFSGVYASGVTPMILTALIAALGGAPWLACGYLVATAVISVLATALI
MAB_2481|M.abscessus_ATCC_1997 LGYQIGAVLSGGFAPLIAAALLAANHGDPRLIVCYYLALSVLTFGAACVA
MSMEG_3398|M.smegmatis_MC2_155 LGYNLAGALS-GFLPLIATALLAVGDGASWPAALVLIGICAITAIGGLAG
MLBr_01222|M.leprae_Br4923 -RDDLGGRLCSR--------------------------------------
..
Mb1232|M.bovis_AF2122/97 PETAG---------SALVSR-------
Rv1200|M.tuberculosis_H37Rv PETAG---------SALVSR-------
MMAR_4238|M.marinum_M PETAG---------APLTAA-------
MUL_0949|M.ulcerans_Agy99 PETAG---------APLTAA-------
Mvan_2899|M.vanbaalenii_PYR-1 SETRG---------TDLTTV-------
Mflv_2383|M.gilvum_PYR-GCK PGGWR---AAPAEARPEHTPTAPTG--
TH_3714|M.thermoresistible__bu PGGWGRTGDQPATEVPASVRPMVSGPD
MAV_4627|M.avium_104 R------------DRDLYL--------
MAB_2481|M.abscessus_ATCC_1997 RETYR---------DQIGDSL------
MSMEG_3398|M.smegmatis_MC2_155 ERMRLR-------HDPVEHQPATS---
MLBr_01222|M.leprae_Br4923 ---------------------------