For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTDAADTPLGELNVSPTGWALLGMLSGGDELSGYDIKKWINWAIQFFYSSPAYSQIYSELKRLERLGLVS SRVDAGARSRRMYKITDAGLSAVTRWANEEAVEPPTLKHNPVLRVMLGHLLKPGRLREVLAEHADYADRM HQAAATEARWTEDQPAWSYAGLALRWSERYYAAERELALQMIKDLDAVEAQLAETGAQNMEFPVREYWYE VERRIAAEETDETPPGAQRPD
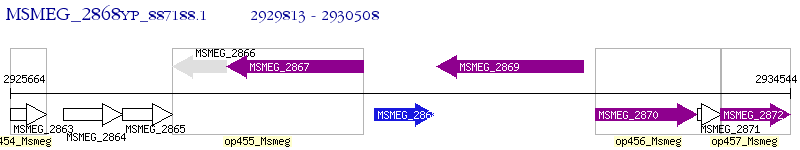
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2868 | - | - | 100% (231) | transcriptional regulator, PadR family protein |
| M. smegmatis MC2 155 | MSMEG_6226 | - | 4e-10 | 28.48% (158) | transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_6605 | - | 4e-07 | 30.25% (119) | transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4568 | - | 3e-68 | 57.59% (224) | PadR-like family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0082c | - | 2e-62 | 51.60% (219) | hypothetical protein MAB_0082c |
| M. marinum M | MMAR_3454 | - | 7e-13 | 33.53% (170) | putative regulatory protein |
| M. avium 104 | MAV_3044 | - | 2e-64 | 53.99% (213) | transcriptional regulator, PadR family protein |
| M. thermoresistible (build 8) | TH_0324 | - | 1e-92 | 73.42% (222) | transcriptional regulator, PadR family protein |
| M. ulcerans Agy99 | MUL_2728 | - | 1e-11 | 32.35% (170) | putative regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_2867 | - | 1e-100 | 78.80% (217) | PadR-like family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2868|M.smegmatis_MC2_155 MTDAADTPLG-ELNVSPTGWALLGMLSGGDELSGYDIKKWINWAIQFFYS
Mvan_2867|M.vanbaalenii_PYR-1 --------------MSPTGWALLGMLSGGEEMSGYDIKKWINWAIRFFYS
TH_0324|M.thermoresistible__bu VTEAVEDAAD-QPNLSPTGWALLGMLSGGNELSGYDIKKWTDWVIRFFYS
Mflv_4568|M.gilvum_PYR-GCK MTDSADSGSGRKPGLAATSYALLGLLSYEQELSGYDIRKWIGWTMRFYYG
MAB_0082c|M.abscessus_ATCC_199 -MAAISSPG--EPNLAATSWTILGMLSYEEEVSGYDIKKWADWSISYFYW
MAV_3044|M.avium_104 -MASETDPASQKPALAATSWALLGMMSYEEEVSGYDLKKWIDWSVDLYYW
MMAR_3454|M.marinum_M ---------------MSLRHAALGLLS-QEPGSGYDLLKRFQLSMNNVWP
MUL_2728|M.ulcerans_Agy99 ---------------MSLRHAALGLLS-QEPGSGYDLLKRFQLSMNNVWP
. : **::* : ****: * : :
MSMEG_2868|M.smegmatis_MC2_155 SPAYSQIYSELKRLERLGLVSSRVD--AGARSRRMYKITDAGLSAVTRWA
Mvan_2867|M.vanbaalenii_PYR-1 SPAYSQIYSELKRLERLGLVSSRVD--AGVRSRRMYKITESGLAAVTRWA
TH_0324|M.thermoresistible__bu SPAYSQIYSELKRLEQYGLVTSRIE--GSTRSRRMYKITDAGLAAVTRWT
Mflv_4568|M.gilvum_PYR-GCK SPAYSQIYSELKKLESLGLVTSRVESTGGARSRRLYKIAPAGLDEVIRWA
MAB_0082c|M.abscessus_ATCC_199 SPSFSQVYSELRKLEKIGYATSRVDHDNGARSRRLYKITPAGMAAMSQWA
MAV_3044|M.avium_104 SPSYSQIYTELKKLEALGLVTSRVERDEGTRSRRLYKITPAGMDAVTQWT
MMAR_3454|M.marinum_M -ATQSQLYSELNKLAAAGLIKSSDL---GPRGRKEYAITDAGRAELYRWM
MUL_2728|M.ulcerans_Agy99 -ATQSQLYSELNKLAAAGLIKSSDL---GPRGRKEYAITDAGRAELYRWM
.: **:*:**.:* * .* . *.*: * *: :* : :*
MSMEG_2868|M.smegmatis_MC2_155 NEEAVEPPTLKHNPVLRVMLGHLLKPGRLREVLAEHADYADR--MHQAAA
Mvan_2867|M.vanbaalenii_PYR-1 NEDPVEAPTLKHNPILRMMLGHLLKPGRLREILAGHAAYAEQ--LQQAAA
TH_0324|M.thermoresistible__bu NEEPVEAPTLKHSPMLRVVFGHLTNPGRLKEVLAEHAAYADR--MQREAA
Mflv_4568|M.gilvum_PYR-GCK NDEPFDPPSLKHGPLLRMTFGHLSTPARLKELLNEHIAYADE--MEREAA
MAB_0082c|M.abscessus_ATCC_199 NETPAEPQVLKHSVVLRVMQGHMTDPARLKEILERHIEYAEN--MQRRAA
MAV_3044|M.avium_104 NHAPVDPPVLKHSVLLRVTFGHLSNPGRLKELLQEHVAYAEA--RHRKAM
MMAR_3454|M.marinum_M TSPQQDPP-VRNAALLRVFLLGQLDPSQARSYLESLLSLSEQEKTHYEQI
MUL_2728|M.ulcerans_Agy99 TSPQQDPP-VRNAALLRVFLLGQLDPSQAGSYLESLLSLSEQEKTHYERI
. :. ::: :**: *.: . * :: .
MSMEG_2868|M.smegmatis_MC2_155 TEARWTEDQPAWSYAGLALRWSERYYAAERELALQMIKDLDAVEAQLAET
Mvan_2867|M.vanbaalenii_PYR-1 TEVRWTSEQPAWSYARLALKWSEQYYAAERELALQMIKDLDAAEEAFGEA
TH_0324|M.thermoresistible__bu TEARHTAEQPAWSYARLALQWSERYYAAERDLALQLIKDLDVADRIFTEA
Mflv_4568|M.gilvum_PYR-GCK KDARLAGADPSWAYARVALRWAERYYANERELALQMIKDLDEAEAEFPRA
MAB_0082c|M.abscessus_ATCC_199 LDAAGAAAQPAWAYSRVALKWAERYYAAERDLAMMMIEDLAEAAEEFDKA
MAV_3044|M.avium_104 EDAEGAEAEPAWAYSVLALRWAAKYYAAEREFALEMIKEIDEADAILQRA
MMAR_3454|M.marinum_M RDAYDWAEDDDSFFARAALEQGLRWAQQEIEWAKWVIAGLDQRSGVAPAG
MUL_2728|M.ulcerans_Agy99 RYAYDWAEDDDSFFARAAPEQGLRWAQQEIEWAKWVIAGLDQRSGVAPAG
. : :: * . . :: * : * :* :
MSMEG_2868|M.smegmatis_MC2_155 GA---QNMEFPVREYWYEVERRIAAEETDETPPGAQRPD-
Mvan_2867|M.vanbaalenii_PYR-1 GR---EGVQFPMPEYWYEVERRIAAEDT----PGVEDPSA
TH_0324|M.thermoresistible__bu EQ---SGLEWPVREYWYEVERRIAAEKD------RE----
Mflv_4568|M.gilvum_PYR-GCK AHGDQAAVAWPEPAYWYEVEQRAQSEADS-----------
MAB_0082c|M.abscessus_ATCC_199 GP-DG-GTPRLVPGYWREVEKRVDAEKLSTQHDTAD----
MAV_3044|M.avium_104 AK-GGYGKPRPTPGYWREVEKQVQAKREAE----------
MMAR_3454|M.marinum_M QG--------------------------------------
MUL_2728|M.ulcerans_Agy99 QG--------------------------------------