For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTGIGGTALKFGVFGVVMTLMTGVLFVIFGEYRSGSANTYSAVFDDVSSLKPGDSVRVAGLRVGTVNSVA LQPDNTVVVKFDADDDVRLTEGTTVKVRYLNLVGDHYLELVDEPGSARIQPPGSQIGSDRTESALDLDLL LGGLKPVIQGLNPQDINALTSSLIQILQGQSGDVEALFARTSAFTNTLADNGQTVEQLIVNLNDVLTTVA RDGEKFSGAVDHLETLVTGLAADRDPIAEAITALDNGTASLASLLTEARPPLSGTVDQMERLAPLLSNDD DLARLDLALQKTPANYRKLVRLGSYGSWLNLYICGVSVRVTDLQGRTAHFPWVIQNTGRCAEP
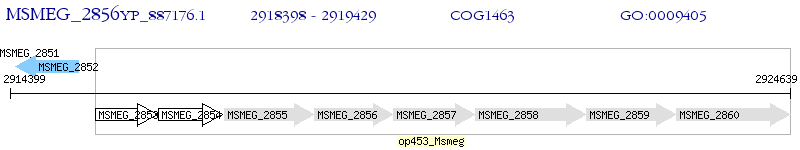
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2856 | - | - | 100% (343) | virulence factor Mce family protein |
| M. smegmatis MC2 155 | MSMEG_4793 | - | e-124 | 65.01% (343) | virulence factor Mce family protein |
| M. smegmatis MC2 155 | MSMEG_5899 | - | 2e-69 | 41.00% (339) | virulence factor Mce family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3528c | mce4B | 6e-68 | 41.67% (336) | MCE-family protein MCE4B |
| M. gilvum PYR-GCK | Mflv_4537 | - | 1e-151 | 76.97% (343) | virulence factor Mce family protein |
| M. tuberculosis H37Rv | Rv3498c | mce4B | 6e-68 | 41.67% (336) | MCE-family protein MCE4B |
| M. leprae Br4923 | MLBr_02590 | mce1B | 8e-58 | 36.26% (342) | putative secreted protein |
| M. abscessus ATCC 19977 | MAB_4152c | - | 6e-67 | 39.94% (338) | MCE-family protein |
| M. marinum M | MMAR_4709 | - | 1e-118 | 62.39% (343) | MCE-family protein |
| M. avium 104 | MAV_0949 | - | 1e-117 | 62.61% (337) | virulence factor Mce family protein |
| M. thermoresistible (build 8) | TH_2568 | - | 1e-164 | 83.09% (343) | PUTATIVE virulence factor Mce family protein |
| M. ulcerans Agy99 | MUL_4061 | mce4B | 7e-70 | 41.96% (336) | MCE-family protein Mce4B |
| M. vanbaalenii PYR-1 | Mvan_2852 | - | 1e-153 | 78.82% (340) | virulence factor Mce family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3528c|M.bovis_AF2122/97 MAGSGVPSHRSMVIKVSVFAVVMLLVAAGLVVVFGDFRFGPTTVYHATFT
Rv3498c|M.tuberculosis_H37Rv MAGSGVPSHRSMVIKVSVFAVVMLLVAAGLVVVFGDFRFGPTTVYHATFT
MUL_4061|M.ulcerans_Agy99 MASGGMPSHRSMVIKVSVFGVVMLVLAAALVVVFGDFRFGPTNVYHATFT
MAB_4152c|M.abscessus_ATCC_199 -------MKRATQIKVIVFTVVMLLVAAGLIITFGEFRFGSGKRYHAMFS
MSMEG_2856|M.smegmatis_MC2_155 -----MTGIGGTALKFGVFGVVMTLMTGVLFVIFGEYRSGSANTYSAVFD
TH_2568|M.thermoresistible__bu -----MTGIGGTAAKFGIFGTVMLLLTGMLFVIFGEYRSGSTTDYSAVFD
Mflv_4537|M.gilvum_PYR-GCK -----MTDVGKAATKFAAFGLTMVVLTAGLFMIFGEYRSGAANRYSAVFT
Mvan_2852|M.vanbaalenii_PYR-1 --------MGGAATKFAAFGLTMVLLTAGLFVIFGEYRSGSTHGYSAVFA
MMAR_4709|M.marinum_M -----MTGSRAMIIKFGAFAAVMIVLTVFLFFIFGQYRTGSTNGYSALFN
MAV_0949|M.avium_104 -----MR-TRATLIKFAIFAVVMAMLTAFLFFIFGQYRTGATNGYSAVFT
MLBr_02590|M.leprae_Br4923 ------MKLTGTVVRLSIFSLVLLLFTVMIIVVFGQMRFGRTNGYTAEFT
:. * .: :.: :.. **: * * * * *
Mb3528c|M.bovis_AF2122/97 DASRLKAGQKVRIAGVPVGSVKAVKLNPD-HSIDVAFAIDRSYTLYSSTR
Rv3498c|M.tuberculosis_H37Rv DASRLKAGQKVRIAGVPVGSVKAVKLNPD-HSIDVAFAIDRSYTLYSSTR
MUL_4061|M.ulcerans_Agy99 NASRLKAGQKVRIAGVPVGSVKAVDLNPD-NTIDVAFAIDRAYTMYSSTR
MAB_4152c|M.abscessus_ATCC_199 TASDLRSGQKVRIAGVPVGRVKNVSLNRD-NSVEVTFDVDSKYQLYSSSR
MSMEG_2856|M.smegmatis_MC2_155 DVSSLKPGDSVRVAGLRVGTVNSVALQPD-NTVVVKFDADDDVRLTEGTT
TH_2568|M.thermoresistible__bu DVSSLKRGDSVRVAGLRVGTVKDVSLQQD-NTVLVRFTADDKVRLTEGTR
Mflv_4537|M.gilvum_PYR-GCK DSSSLTSGDSVRAAGIRVGTVRGVTLQDD-NTVVVDFDADDNVRLTEGTR
Mvan_2852|M.vanbaalenii_PYR-1 DSSSLESGDSVRVAGIRIGTVRSVNLQAD-NTVVVDFDADDDIRLTESTK
MMAR_4709|M.marinum_M DVSRLKPGETVRIAGVRVGTVNSVSLRTD-KKVLVKFDADRNIVLTTGTR
MAV_0949|M.avium_104 DVSRLKPGQSVRVAGIRVGTVNSVSLQPD-KKVVVKFDADRNIVLTEGTR
MLBr_02590|M.leprae_Br4923 NISGLRTGQFVRASGVEVGKVNSVALINGGTRVRVKFNVDRSVPLYQSTT
* * *: ** :*: :* *. * * . : * * * : .:
Mb3528c|M.bovis_AF2122/97 AVIRYENLVGDRFLEITSG--PGELRKLPPGGTINVAHTQPALDLDALLG
Rv3498c|M.tuberculosis_H37Rv AVIRYENLVGDRFLEITSG--PGELRKLPPGGTINVAHTQPALDLDALLG
MUL_4061|M.ulcerans_Agy99 AVIRYENLVGDRYLEITSG--PDELRKLPPGGTINAEHTQPALDLDALLG
MAB_4152c|M.abscessus_ATCC_199 AIIRYENLVGDRYLEITAG--PGELHKLPDGGTLDKEHTQPALDLDALLG
MSMEG_2856|M.smegmatis_MC2_155 VKVRYLNLVGDHYLELVDE--PGSARIQPPGSQIGSDRTESALDLDLLLG
TH_2568|M.thermoresistible__bu AKVRYLNLVGDRYLELVDE--PGSTRIQPPGSLIGSDRTEPALDLDLLLG
Mflv_4537|M.gilvum_PYR-GCK IAVRYLNLVGDRYLELLDE--PGSARIQPSGSRIGLERTEPALNLDLLLG
Mvan_2852|M.vanbaalenii_PYR-1 VAVRYLNLVGDRYLELLDA--PGSTKLQRPGSRIGVERTEPALNLDLLLG
MMAR_4709|M.marinum_M AVVRYLNLVGDRYLELVDG--PGSTKTLPAGSQIPIERTAGALDLDLLLG
MAV_0949|M.avium_104 AAVRYLNLVGDRYLELVDG--PGSPKRLPAGGQIPVSRTAPALDLDLLLG
MLBr_02590|M.leprae_Br4923 AQIRYLDLIGNRYLELKHGEGQGSEKIMPAGGLIPLSRTSPALDLDALIG
:** :*:*:::**: .. : *. : :* **:** *:*
Mb3528c|M.bovis_AF2122/97 GLRPVLKGFDADKINTITSAVIELLQGQGGPLANVLADTGAFSAALGARD
Rv3498c|M.tuberculosis_H37Rv GLRPVLKGFDADKINTITSAVIELLQGQGGPLANVLADTGAFSAALGARD
MUL_4061|M.ulcerans_Agy99 GLRPVLKGLDADKVNTVTGAVIELLQGQGGALANVLADTSAFSTALARRD
MAB_4152c|M.abscessus_ATCC_199 GLRPVLKGFDAAKVNEISNAFIQILQGQGGTLSQLLGDTSSFTSGLAERD
MSMEG_2856|M.smegmatis_MC2_155 GLKPVIQGLNPQDINALTSSLIQILQGQSGDVEALFARTSAFTNTLADNG
TH_2568|M.thermoresistible__bu GLKPVIQGLNPQDVNALTGALIQALQGQGGDIESLFSHTSSFTNTLADNG
Mflv_4537|M.gilvum_PYR-GCK GLKPVIQGLNPDDVNALTSSLIQILQGQGGNLESLFSRTSSFTNSLADNG
Mvan_2852|M.vanbaalenii_PYR-1 GLKPVIQGLNPDDVNALTSSLIQILQGQGGNLESLFARTASFTNALADNG
MMAR_4709|M.marinum_M GLKPVTQGLNPQDVNALSASLIQIFQGEGSTLESLLSKTSSFTNALADNN
MAV_0949|M.avium_104 GLKPVTQGLNARDVNALTSGLLQVFQGQGGTLDSLFTKATSFSNALADND
MLBr_02590|M.leprae_Br4923 GFKPLFRALDPDKINTIASAIVAAFQGQGGTINDILDQTAQLTTAIGERD
*::*: :.::. .:* :: ..: :**:.. : :: : :: :. ..
Mb3528c|M.bovis_AF2122/97 QLIGEVITNLNAVLATVDAKSAQFSASVDQLQQLVSGLAKNRDPIAGAIS
Rv3498c|M.tuberculosis_H37Rv QLIGEVITNLNAVLATVDAKSAQFSASVDQLQQLVSGLAKNRDPIAGAIS
MUL_4061|M.ulcerans_Agy99 QLIGDTITNLNSVLATVDQRSAQFSASVDQLQQLITGLAENKDVIAGAIP
MAB_4152c|M.abscessus_ATCC_199 QLIGDVINHLNEVLATVDSKSTQFSSSVDQLQQLISGLAQQRDVVAGALP
MSMEG_2856|M.smegmatis_MC2_155 QTVEQLIVNLNDVLTTVARDGEKFSGAVDHLETLVTGLAADRDPIAEAIT
TH_2568|M.thermoresistible__bu RVVEQLIDNLNEVLNTIAEDGDKFSGAVNHLENLITRLAADRDPIGEAIT
Mflv_4537|M.gilvum_PYR-GCK QTVERLIDTLNETLATVSADGQNLSSAIDQLEQLATGLAQDRDPIGDAIT
Mvan_2852|M.vanbaalenii_PYR-1 QTVERLIETLNETIGTVAADGEKFSGAIDQLEQLATGLAQDRDPIGEAIT
MMAR_4709|M.marinum_M ETVQALIDNLNVVVDTVNREGTKFSGAIDRLERLISALSQDRDTIGAAIT
MAV_0949|M.avium_104 QTVQQLIDNLNIVIGTISKDGKQFSGAVDRLERLVSGLSDDRNTIGSAID
MLBr_02590|M.leprae_Br4923 QAIGEVVKNLNVVLDTTVRHRKEFDQTVNNLEILITGLKDHGDQLAGSFA
. : : ** .: * ::. :::.*: * : * . : :. ::
Mb3528c|M.bovis_AF2122/97 PLASTTTDLTELLRNSRRPLQGILENARPLATELDNRKAE--VNNDIEQL
Rv3498c|M.tuberculosis_H37Rv PLASTTTDLTELLRNSRRPLQGILENARPLATELDNRKAE--VNNDIEQL
MUL_4061|M.ulcerans_Agy99 PLASTTTDLTELLKNSRRPLQGLLENARPLATELDNRKDE--VNNDVEQL
MAB_4152c|M.abscessus_ATCC_199 PLASTASSLESVLKDTRPYIKGTIEQARQLATRVDERKAD--VNVVVENL
MSMEG_2856|M.smegmatis_MC2_155 ALDNGTASLASLLTEARPPLSGTVDQMERLAPLLSNDDDLARLDLALQKT
TH_2568|M.thermoresistible__bu ALDNGTASLADLLGRSRPPLAGTVDELARLAPLLSNDTDLARIDLALQKT
Mflv_4537|M.gilvum_PYR-GCK ALDSGTASLAGLLTEARPPLAGTVDQLNRLAPLLSNETDLARLDLALQKT
Mvan_2852|M.vanbaalenii_PYR-1 ALDNGTASLAGLLTEARPPLAGTVDQLNRLAPLLSNETDLARLDLALQKT
MMAR_4709|M.marinum_M ALDNGTTSIADLLGRARAPLAGTVDQLNRLAPLLDKDKNL--IDISLQKL
MAV_0949|M.avium_104 ALDRGTASLADLLAQARPPLTGTIDQLNRLAPILDNDKDR--LDAAIGKA
MLBr_02590|M.leprae_Br4923 HISNAAGTVANLLAEDHALLHKSINYLDGIQQPLIDQRLQ--LEDYLHKL
: : : :* : : :: : : . :: : :
Mb3528c|M.bovis_AF2122/97 GEDYLRLSA-LGSYGAFFNIYFCSVTIKINGPAGSDILLPIGGQPDPSKG
Rv3498c|M.tuberculosis_H37Rv GEDYLRLSA-LGSYGAFFNIYFCSVTIKINGPAGSDILLPIGGQPDPSKG
MUL_4061|M.ulcerans_Agy99 GEDYLRLAA-LGSYGAFFNIYFCTVTIKINGPAGSDILIPMGGQPDPSKG
MAB_4152c|M.abscessus_ATCC_199 AENYLRLNA-LGAYGSFFNIYYCSIRIKVNGPVGTDWLIPFGGPPDPSKG
MSMEG_2856|M.smegmatis_MC2_155 PANYRKLVR-LGSYGSWLNLYICGVSVRVTDLQGRTAHFPWVIQ---NTG
TH_2568|M.thermoresistible__bu PANYRKLVR-LGAYGSWLNLYICGVSVRVTDLQGRTAHFPWVIQ---NTG
Mflv_4537|M.gilvum_PYR-GCK PQNYRKLVR-LGSYGSWLNLYICGVSIRVSDLQGRTAHFPWVIQ---NTG
Mvan_2852|M.vanbaalenii_PYR-1 PQNYRKLVR-LGSYGSWLNLYICGVSIRVSDLQGRTAHFPWVIQ---NTG
MMAR_4709|M.marinum_M PDNYRKLTR-LGSYGAWFPYYLCGLALRVSDLQYRTVEVGITHQ---VTG
MAV_0949|M.avium_104 PKNYRKLVR-LGANGATIPYYLCMLELRGTDLQGKTVRAPIFRS---DAG
MLBr_02590|M.leprae_Br4923 PTVLNQIGRTIGSYGDFVNFYSCDITLKINGLQ-PGGPVRTVRLFQQPTG
:: :*: * . * * : :: .. *
Mb3528c|M.bovis_AF2122/97 RCAFAK-
Rv3498c|M.tuberculosis_H37Rv RCAFAK-
MUL_4061|M.ulcerans_Agy99 RCAFAK-
MAB_4152c|M.abscessus_ATCC_199 RCAFKNE
MSMEG_2856|M.smegmatis_MC2_155 RCAEP--
TH_2568|M.thermoresistible__bu RCAEP--
Mflv_4537|M.gilvum_PYR-GCK RCGEP--
Mvan_2852|M.vanbaalenii_PYR-1 RCAEP--
MMAR_4709|M.marinum_M RCAEPDA
MAV_0949|M.avium_104 RCTEP--
MLBr_02590|M.leprae_Br4923 RCTPQ--
**