For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRTRATLIKFAIFAVVMAMLTAFLFFIFGQYRTGATNGYSAVFTDVSRLKPGQSVRVAGIRVGTVNSVSL QPDKKVVVKFDADRNIVLTEGTRAAVRYLNLVGDRYLELVDGPGSPKRLPAGGQIPVSRTAPALDLDLLL GGLKPVTQGLNARDVNALTSGLLQVFQGQGGTLDSLFTKATSFSNALADNDQTVQQLIDNLNIVIGTISK DGKQFSGAVDRLERLVSGLSDDRNTIGSAIDALDRGTASLADLLAQARPPLTGTIDQLNRLAPILDNDKD RLDAAIGKAPKNYRKLVRLGANGATIPYYLCMLELRGTDLQGKTVRAPIFRSDAGRCTEP
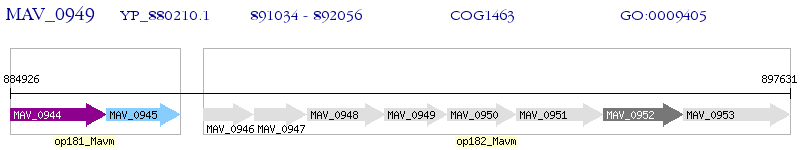
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0949 | - | - | 100% (340) | virulence factor Mce family protein |
| M. avium 104 | MAV_0658 | - | 4e-72 | 41.25% (337) | virulence factor Mce family protein |
| M. avium 104 | MAV_2062 | - | 1e-71 | 41.21% (330) | virulence factor Mce family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3528c | mce4B | 2e-74 | 43.87% (326) | MCE-family protein MCE4B |
| M. gilvum PYR-GCK | Mflv_2509 | - | 1e-121 | 61.54% (338) | virulence factor Mce family protein |
| M. tuberculosis H37Rv | Rv3498c | mce4B | 2e-74 | 43.87% (326) | MCE-family protein MCE4B |
| M. leprae Br4923 | MLBr_02590 | mce1B | 8e-65 | 36.92% (344) | putative secreted protein |
| M. abscessus ATCC 19977 | MAB_4152c | - | 1e-69 | 42.06% (321) | MCE-family protein |
| M. marinum M | MMAR_4709 | - | 1e-137 | 70.41% (338) | MCE-family protein |
| M. smegmatis MC2 155 | MSMEG_4793 | - | 1e-125 | 64.60% (339) | virulence factor Mce family protein |
| M. thermoresistible (build 8) | TH_4265 | - | 1e-123 | 64.18% (335) | PUTATIVE virulence factor Mce family protein |
| M. ulcerans Agy99 | MUL_4061 | mce4B | 8e-74 | 42.64% (326) | MCE-family protein Mce4B |
| M. vanbaalenii PYR-1 | Mvan_4149 | - | 1e-124 | 63.31% (338) | virulence factor Mce family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3528c|M.bovis_AF2122/97 MAGSGVPSHRSMVIKVSVFAVVMLLVAAGLVVVFGDFRFGPTTVYHATFT
Rv3498c|M.tuberculosis_H37Rv MAGSGVPSHRSMVIKVSVFAVVMLLVAAGLVVVFGDFRFGPTTVYHATFT
MUL_4061|M.ulcerans_Agy99 MASGGMPSHRSMVIKVSVFGVVMLVLAAALVVVFGDFRFGPTNVYHATFT
MAB_4152c|M.abscessus_ATCC_199 -------MKRATQIKVIVFTVVMLLVAAGLIITFGEFRFGSGKRYHAMFS
MAV_0949|M.avium_104 -----MR-TRATLIKFAIFAVVMAMLTAFLFFIFGQYRTGATNGYSAVFT
MMAR_4709|M.marinum_M -----MTGSRAMIIKFGAFAAVMIVLTVFLFFIFGQYRTGSTNGYSALFN
MSMEG_4793|M.smegmatis_MC2_155 -----MRRASSTLVKFGVFAVVMAVLTAFLFMTFSEYRGGSYAGYSAVFD
TH_4265|M.thermoresistible__bu --------MTRTIVKFGVFVTVMTTLTAFLFMTFSQYRGGATSRYSAVFE
Mflv_2509|M.gilvum_PYR-GCK -----MTRSTATLIKFTVFGLVMAVLTAFLFLVFSDTRTGAANEYSAVFK
Mvan_4149|M.vanbaalenii_PYR-1 -----MTRSAATLVKFTAFGLVMALLTAFLFLVFSDTRTGAANEYTAVFK
MLBr_02590|M.leprae_Br4923 ------MKLTGTVVRLSIFSLVLLLFTVMIIVVFGQMRFGRTNGYTAEFT
::. * *: .:. :.. *.: * * * * *
Mb3528c|M.bovis_AF2122/97 DASRLKAGQKVRIAGVPVGSVKAVKLNPD-HSIDVAFAIDRSYTLYSSTR
Rv3498c|M.tuberculosis_H37Rv DASRLKAGQKVRIAGVPVGSVKAVKLNPD-HSIDVAFAIDRSYTLYSSTR
MUL_4061|M.ulcerans_Agy99 NASRLKAGQKVRIAGVPVGSVKAVDLNPD-NTIDVAFAIDRAYTMYSSTR
MAB_4152c|M.abscessus_ATCC_199 TASDLRSGQKVRIAGVPVGRVKNVSLNRD-NSVEVTFDVDSKYQLYSSSR
MAV_0949|M.avium_104 DVSRLKPGQSVRVAGIRVGTVNSVSLQPD-KKVVVKFDADRNIVLTEGTR
MMAR_4709|M.marinum_M DVSRLKPGETVRIAGVRVGTVNSVSLRTD-KKVLVKFDADRNIVLTTGTR
MSMEG_4793|M.smegmatis_MC2_155 DASRLESGDSVRVAGVRVGTVKNVSLQPD-RTVLVDFDTERNIALTTGTK
TH_4265|M.thermoresistible__bu DASRLESGDSVRVAGVRVGTVRGVALQPD-KTVLVDFDADRGIQLTEGTR
Mflv_2509|M.gilvum_PYR-GCK DASRLKSGDTVRIAGIRVGTVKDVELQAD-KSVLVTFDADRSTQLTTGTN
Mvan_4149|M.vanbaalenii_PYR-1 DASRLKTGDTVRIAGIRVGTVKDVELQAD-RSVLVTFDADRNTVLTTGTN
MLBr_02590|M.leprae_Br4923 NISGLRTGQFVRASGVEVGKVNSVALINGGTRVRVKFNVDRSVPLYQSTT
* *..*: ** :*: ** *. * * . : * * : : .:
Mb3528c|M.bovis_AF2122/97 AVIRYENLVGDRFLEITSG--PGELRKLPPGGTINVAHTQPALDLDALLG
Rv3498c|M.tuberculosis_H37Rv AVIRYENLVGDRFLEITSG--PGELRKLPPGGTINVAHTQPALDLDALLG
MUL_4061|M.ulcerans_Agy99 AVIRYENLVGDRYLEITSG--PDELRKLPPGGTINAEHTQPALDLDALLG
MAB_4152c|M.abscessus_ATCC_199 AIIRYENLVGDRYLEITAG--PGELHKLPDGGTLDKEHTQPALDLDALLG
MAV_0949|M.avium_104 AAVRYLNLVGDRYLELVDG--PGSPKRLPAGGQIPVSRTAPALDLDLLLG
MMAR_4709|M.marinum_M AVVRYLNLVGDRYLELVDG--PGSTKTLPAGSQIPIERTAGALDLDLLLG
MSMEG_4793|M.smegmatis_MC2_155 AAVRYLNLVGDRYLELIDS--PGSTRLLPAGSQIPKERTASALDLDLLLG
TH_4265|M.thermoresistible__bu AAVRYLNLTGDRYLELLDE--PGSTRLLPAGSRIPKERTAPALDLDLLLG
Mflv_2509|M.gilvum_PYR-GCK VAIRYLNLVGDRYMELVDT--PDSNQILPAGSRIPEDRTAPALDLDVLLG
Mvan_4149|M.vanbaalenii_PYR-1 AAIRYLNLVGDRYLELVDT--PDSTQIVPAGGQIPEDRTTPALDLDVLLG
MLBr_02590|M.leprae_Br4923 AQIRYLDLIGNRYLELKHGEGQGSEKIMPAGGLIPLSRTSPALDLDALIG
. :** :* *:*::*: .. : :* *. : :* ***** *:*
Mb3528c|M.bovis_AF2122/97 GLRPVLKGFDADKINTITSAVIELLQGQGGPLANVLADTGAFSAALGARD
Rv3498c|M.tuberculosis_H37Rv GLRPVLKGFDADKINTITSAVIELLQGQGGPLANVLADTGAFSAALGARD
MUL_4061|M.ulcerans_Agy99 GLRPVLKGLDADKVNTVTGAVIELLQGQGGALANVLADTSAFSTALARRD
MAB_4152c|M.abscessus_ATCC_199 GLRPVLKGFDAAKVNEISNAFIQILQGQGGTLSQLLGDTSSFTSGLAERD
MAV_0949|M.avium_104 GLKPVTQGLNARDVNALTSGLLQVFQGQGGTLDSLFTKATSFSNALADND
MMAR_4709|M.marinum_M GLKPVTQGLNPQDVNALSASLIQIFQGEGSTLESLLSKTSSFTNALADNN
MSMEG_4793|M.smegmatis_MC2_155 GLKPVIQGLDPQDVNALTSSLIQIFQGQGDTLSSLMSKTSSFSNTLADND
TH_4265|M.thermoresistible__bu GLRPVINGLNPQDVNALTSALVQIFQGQGDTLDSLMSKTSSFSTSLADSS
Mflv_2509|M.gilvum_PYR-GCK GLRPVIQGLNPQDVNALTSSLIQILQGQGGTLDSLFAKTSSFTTSLADNN
Mvan_4149|M.vanbaalenii_PYR-1 GLKPVIQGLNPEDVNGLTSALIQILQGQGGTLDSLFSKTSSFSNSLADNN
MLBr_02590|M.leprae_Br4923 GFKPLFRALDPDKINTIASAIVAAFQGQGGTINDILDQTAQLTTAIGERD
*::*: ..::. .:* :: ..: :**:*..: .:: .: :: :. .
Mb3528c|M.bovis_AF2122/97 QLIGEVITNLNAVLATVDAKSAQFSASVDQLQQLVSGLAKNRDPIAGAIS
Rv3498c|M.tuberculosis_H37Rv QLIGEVITNLNAVLATVDAKSAQFSASVDQLQQLVSGLAKNRDPIAGAIS
MUL_4061|M.ulcerans_Agy99 QLIGDTITNLNSVLATVDQRSAQFSASVDQLQQLITGLAENKDVIAGAIP
MAB_4152c|M.abscessus_ATCC_199 QLIGDVINHLNEVLATVDSKSTQFSSSVDQLQQLISGLAQQRDVVAGALP
MAV_0949|M.avium_104 QTVQQLIDNLNIVIGTISKDGKQFSGAVDRLERLVSGLSDDRNTIGSAID
MMAR_4709|M.marinum_M ETVQALIDNLNVVVDTVNREGTKFSGAIDRLERLISALSQDRDTIGAAIT
MSMEG_4793|M.smegmatis_MC2_155 QVIGQLIENLDTVVDTLNRDGGKFSATVDRLEQLISGLSQDRDPIGTAVT
TH_4265|M.thermoresistible__bu QAIQQLIDNLRAVVDTLEKDGDKFSGAIDQLEQLVSGLAEDRDPIGTAIT
Mflv_2509|M.gilvum_PYR-GCK QVIEQLIDDLRTVLDTLAEDGDEFSGAIDKLDQLVGGLAEDRDPIGTAIT
Mvan_4149|M.vanbaalenii_PYR-1 QVIEELIVDLRTVLDTLSKDGEEFSGAIDKLEQLVSGLSSDRDPIGTAIT
MLBr_02590|M.leprae_Br4923 QAIGEVVKNLNVVLDTTVRHRKEFDQTVNNLEILITGLKDHGDQLAGSFA
: : : .* *: * :*. :::.*: *: .* .. : :. :.
Mb3528c|M.bovis_AF2122/97 PLASTTTDLTELLRNSRRPLQGILENARPLATELDNRKAEVNNDIEQLGE
Rv3498c|M.tuberculosis_H37Rv PLASTTTDLTELLRNSRRPLQGILENARPLATELDNRKAEVNNDIEQLGE
MUL_4061|M.ulcerans_Agy99 PLASTTTDLTELLKNSRRPLQGLLENARPLATELDNRKDEVNNDVEQLGE
MAB_4152c|M.abscessus_ATCC_199 PLASTASSLESVLKDTRPYIKGTIEQARQLATRVDERKADVNVVVENLAE
MAV_0949|M.avium_104 ALDRGTASLADLLAQARPPLTGTIDQLNRLAPILDNDKDRLDAAIGKAPK
MMAR_4709|M.marinum_M ALDNGTTSIADLLGRARAPLAGTVDQLNRLAPLLDKDKNLIDISLQKLPD
MSMEG_4793|M.smegmatis_MC2_155 QLDNGTASIATLLTQARPPLDATVDQLNRMAPLLDDHKVVLDRGLQKAPA
TH_4265|M.thermoresistible__bu ALDQGTASMADLLSQARPPLDRTVNELNRLAPLLADHQIVFDRALQRGPE
Mflv_2509|M.gilvum_PYR-GCK ALDNGTASIADLLSRGRAPLSSTVDELSRLAPLVDNDLDRLDATLQRLPE
Mvan_4149|M.vanbaalenii_PYR-1 ALDNGTASIADLLGRGRAPLANTVDEMNRLAPLVDNDLDRLDATLQRLPE
MLBr_02590|M.leprae_Br4923 HISNAAGTVANLLAEDHALLHKSINYLDGIQQPLIDQRLQLEDYLHKLPT
: : : :* : : :: : : . .: : .
Mb3528c|M.bovis_AF2122/97 DYLRLSA-LGSYGAFFNIYFCSVTIKINGPAGSDILLPIGGQPDPSKGRC
Rv3498c|M.tuberculosis_H37Rv DYLRLSA-LGSYGAFFNIYFCSVTIKINGPAGSDILLPIGGQPDPSKGRC
MUL_4061|M.ulcerans_Agy99 DYLRLAA-LGSYGAFFNIYFCTVTIKINGPAGSDILIPMGGQPDPSKGRC
MAB_4152c|M.abscessus_ATCC_199 NYLRLNA-LGAYGSFFNIYYCSIRIKVNGPVGTDWLIPFGGPPDPSKGRC
MAV_0949|M.avium_104 NYRKLVR-LGANGATIPYYLCMLELRGTDLQGKTVRAPIFRS---DAGRC
MMAR_4709|M.marinum_M NYRKLTR-LGSYGAWFPYYLCGLALRVSDLQYRTVEVGITHQ---VTGRC
MSMEG_4793|M.smegmatis_MC2_155 NYRKLAR-LGSYGSWIMYYVCGISIRVTDLQGRTAVFPMIKQ---EGGRC
TH_4265|M.thermoresistible__bu NYRKLAR-LGSYGSWIMYYICGISFRVTDLEGRTAVFPMVKQ---EGGRC
Mflv_2509|M.gilvum_PYR-GCK IYRKLAR-VGSYGAWFPYYICGISFRASDLEGRTVVFPWIKQ---EEGRC
Mvan_4149|M.vanbaalenii_PYR-1 IYRKLAR-VGSYGAWFPYYICGISFRASDLEGRTVVFPWIKQ---EEGRC
MLBr_02590|M.leprae_Br4923 VLNQIGRTIGSYGDFVNFYSCDITLKINGLQ-PGGPVRTVRLFQQPTGRC
:: :*: * . * * : :: .. ***
Mb3528c|M.bovis_AF2122/97 AFAK-
Rv3498c|M.tuberculosis_H37Rv AFAK-
MUL_4061|M.ulcerans_Agy99 AFAK-
MAB_4152c|M.abscessus_ATCC_199 AFKNE
MAV_0949|M.avium_104 TEP--
MMAR_4709|M.marinum_M AEPDA
MSMEG_4793|M.smegmatis_MC2_155 AEP--
TH_4265|M.thermoresistible__bu AEP--
Mflv_2509|M.gilvum_PYR-GCK VDE--
Mvan_4149|M.vanbaalenii_PYR-1 VDE--
MLBr_02590|M.leprae_Br4923 TPQ--
.