For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KRATQIKVIVFTVVMLLVAAGLIITFGEFRFGSGKRYHAMFSTASDLRSGQKVRIAGVPVGRVKNVSLNR DNSVEVTFDVDSKYQLYSSSRAIIRYENLVGDRYLEITAGPGELHKLPDGGTLDKEHTQPALDLDALLGG LRPVLKGFDAAKVNEISNAFIQILQGQGGTLSQLLGDTSSFTSGLAERDQLIGDVINHLNEVLATVDSKS TQFSSSVDQLQQLISGLAQQRDVVAGALPPLASTASSLESVLKDTRPYIKGTIEQARQLATRVDERKADV NVVVENLAENYLRLNALGAYGSFFNIYYCSIRIKVNGPVGTDWLIPFGGPPDPSKGRCAFKNE*
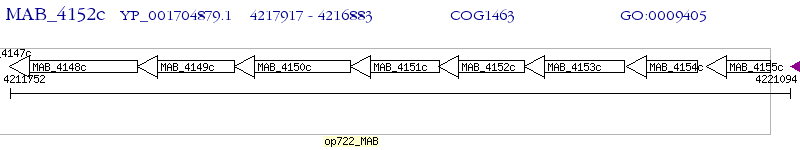
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4152c | - | - | 100% (344) | MCE-family protein |
| M. abscessus ATCC 19977 | MAB_4598c | - | 6e-35 | 28.88% (329) | putative Mce family protein |
| M. abscessus ATCC 19977 | MAB_4515c | - | 4e-34 | 28.16% (316) | putative Mce family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3528c | mce4B | 1e-129 | 65.78% (339) | MCE-family protein MCE4B |
| M. gilvum PYR-GCK | Mflv_1557 | - | 1e-126 | 64.90% (339) | virulence factor Mce family protein |
| M. tuberculosis H37Rv | Rv3498c | mce4B | 1e-129 | 65.78% (339) | MCE-family protein MCE4B |
| M. leprae Br4923 | MLBr_02590 | mce1B | 2e-57 | 36.16% (318) | putative secreted protein |
| M. marinum M | MMAR_4986 | mce4B | 1e-129 | 66.08% (339) | MCE-family protein Mce4B |
| M. avium 104 | MAV_0658 | - | 1e-131 | 67.26% (339) | virulence factor Mce family protein |
| M. smegmatis MC2 155 | MSMEG_5899 | - | 1e-133 | 68.45% (336) | virulence factor Mce family protein |
| M. thermoresistible (build 8) | TH_1936 | - | 1e-128 | 66.17% (334) | MCE-FAMILY PROTEIN MCE4B |
| M. ulcerans Agy99 | MUL_4061 | mce4B | 1e-128 | 65.49% (339) | MCE-family protein Mce4B |
| M. vanbaalenii PYR-1 | Mvan_5201 | - | 1e-128 | 65.29% (340) | virulence factor Mce family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1557|M.gilvum_PYR-GCK -----MNRERRTLVSVSIFTVVMLMVAASLIVVFGEFRFAAEDKYRATFT
Mvan_5201|M.vanbaalenii_PYR-1 -----MR--DKTLINVGVFTVAMLLVAAMLVVVFGEFRFASEDKYHATFT
MSMEG_5899|M.smegmatis_MC2_155 -----MHR-DRTMLKVGIFTMVMLLVAAGLVVVFGEFRFAAGQSYHATFS
TH_1936|M.thermoresistible__bu ------------MIKVVVFTIAMVLVAAGLVVVFGEFRFASGRTYHATFT
MAB_4152c|M.abscessus_ATCC_199 -----MKR--ATQIKVIVFTVVMLLVAAGLIITFGEFRFGSGKRYHAMFS
Mb3528c|M.bovis_AF2122/97 MAGSGVPSHRSMVIKVSVFAVVMLLVAAGLVVVFGDFRFGPTTVYHATFT
Rv3498c|M.tuberculosis_H37Rv MAGSGVPSHRSMVIKVSVFAVVMLLVAAGLVVVFGDFRFGPTTVYHATFT
MMAR_4986|M.marinum_M MASGGMPSHRSMVIKVSVFGVVMLVVAAALVVVFGDFRFGPTNVYHATFT
MUL_4061|M.ulcerans_Agy99 MASGGMPSHRSMVIKVSVFGVVMLVLAAALVVVFGDFRFGPTNVYHATFT
MAV_0658|M.avium_104 MAGSGMPSHRSMVIKVSIFAAAMLLVSAGLVVVFGDFRFGPESTYHATFV
MLBr_02590|M.leprae_Br4923 ------MKLTGTVVRLSIFSLVLLLFTVMIIVVFGQMRFGRTNGYTAEFT
: : :* .:::.:. :::.**::**. * * *
Mflv_1557|M.gilvum_PYR-GCK DASRLKTGQDVRIAGIPVGTVEGLSLNPDNS-VEVEFTVNERYQLYTSTK
Mvan_5201|M.vanbaalenii_PYR-1 DASRLKAGQDVRIAGVPVGTVEGLTLNPDNT-VDVAFTVNDRYQLYTSTK
MSMEG_5899|M.smegmatis_MC2_155 EASRLKAGQDVRIAGVPVGTVKSVKLNPDNT-VDVGFDVNKRYQLYTSSR
TH_1936|M.thermoresistible__bu DASRLETGDDVRIAGVPVGTVKSVKLNEDNT-VDVSFDVNKRYQLYTSTR
MAB_4152c|M.abscessus_ATCC_199 TASDLRSGQKVRIAGVPVGRVKNVSLNRDNS-VEVTFDVDSKYQLYSSSR
Mb3528c|M.bovis_AF2122/97 DASRLKAGQKVRIAGVPVGSVKAVKLNPDHS-IDVAFAIDRSYTLYSSTR
Rv3498c|M.tuberculosis_H37Rv DASRLKAGQKVRIAGVPVGSVKAVKLNPDHS-IDVAFAIDRSYTLYSSTR
MMAR_4986|M.marinum_M NASRLKAGQKVRIAGVPVGSVKAVDLNPDNT-IDVAFAIDRAYTMYSSTR
MUL_4061|M.ulcerans_Agy99 NASRLKAGQKVRIAGVPVGSVKAVDLNPDNT-IDVAFAIDRAYTMYSSTR
MAV_0658|M.avium_104 DVSRLKAGQKVRIAGVPVGAVEAVKLNRDNT-IDVTFGVDKRYTLYSSTR
MLBr_02590|M.leprae_Br4923 NISGLRTGQFVRASGVEVGKVNSVALINGGTRVRVKFNVDRSVPLYQSTT
* *.:*: ** :*: ** *: : * . : : * * :: :* *:
Mflv_1557|M.gilvum_PYR-GCK AVIRYENLVGDRYLEITSGPGD--LVKLPPGGTV--VNTEPALDLDALLG
Mvan_5201|M.vanbaalenii_PYR-1 AVVRYENLVGDRYLEITSGPGD--LVKLPKGGTI--THTEPALDLDALLG
MSMEG_5899|M.smegmatis_MC2_155 AVVRYENLVGDRYLEITSGPGE--LRKLAPGGTLGLQNTQPALDLDALLG
TH_1936|M.thermoresistible__bu ALIRYENLVGDRYLEIASGPGE--LKKLPPGGRIPQTNTQPALDLDALLG
MAB_4152c|M.abscessus_ATCC_199 AIIRYENLVGDRYLEITAGPGE--LHKLPDGGTLDKEHTQPALDLDALLG
Mb3528c|M.bovis_AF2122/97 AVIRYENLVGDRFLEITSGPGE--LRKLPPGGTINVAHTQPALDLDALLG
Rv3498c|M.tuberculosis_H37Rv AVIRYENLVGDRFLEITSGPGE--LRKLPPGGTINVAHTQPALDLDALLG
MMAR_4986|M.marinum_M AVIRYENLVGDRYLEITSGPGE--LRKLPPGGTINAEHTQPALDLDALLG
MUL_4061|M.ulcerans_Agy99 AVIRYENLVGDRYLEITSGPDE--LRKLPPGGTINAEHTQPALDLDALLG
MAV_0658|M.avium_104 AVIRYENLVGDRYLEITSGPGE--LRKLPAGGTINSQHTQPALDLDALLG
MLBr_02590|M.leprae_Br4923 AQIRYLDLIGNRYLELKHGEGQGSEKIMPAGGLIPLSRTSPALDLDALIG
* :** :*:*:*:**: * .: :. ** : .*.********:*
Mflv_1557|M.gilvum_PYR-GCK GLKPVVKGLDGAKVNEISSAVIELLQGEGGALAAMLSSTSAFTQNLAARD
Mvan_5201|M.vanbaalenii_PYR-1 GLKPVVKGLDGAKINEISNAVIELLQGEGGALSAMLSNTSAFTQNLAARD
MSMEG_5899|M.smegmatis_MC2_155 GLRPVLKGLDGAKINEVSNAVIEMLQGQGGALANMLTSTSAFTQNLAARD
TH_1936|M.thermoresistible__bu GLRPVLKGLEGHKVNEISTAVIQALQGQGGALSELLANTGSFTQRLADRY
MAB_4152c|M.abscessus_ATCC_199 GLRPVLKGFDAAKVNEISNAFIQILQGQGGTLSQLLGDTSSFTSGLAERD
Mb3528c|M.bovis_AF2122/97 GLRPVLKGFDADKINTITSAVIELLQGQGGPLANVLADTGAFSAALGARD
Rv3498c|M.tuberculosis_H37Rv GLRPVLKGFDADKINTITSAVIELLQGQGGPLANVLADTGAFSAALGARD
MMAR_4986|M.marinum_M GLRPVLKGLDADKVNTVTGAVIELLQGQGGALANVLADTSAFSTALARRD
MUL_4061|M.ulcerans_Agy99 GLRPVLKGLDADKVNTVTGAVIELLQGQGGALANVLADTSAFSTALARRD
MAV_0658|M.avium_104 GLRPVLKGLDADKVNTISSAVIQLLQGQGGALSNVLADTSAFTSALGQRD
MLBr_02590|M.leprae_Br4923 GFKPLFRALDPDKINTIASAIVAAFQGQGGTINDILDQTAQLTTAIGERD
*::*:.:.:: *:* :: *.: :**:**.: :* .*. :: :. *
Mflv_1557|M.gilvum_PYR-GCK QLIGDVITNLNTVLGTVDEKGAEFDASVDQLQKLITGLAEGRDPIAGAIG
Mvan_5201|M.vanbaalenii_PYR-1 QLIGDVINNLNTVLGTVDEKGAEFNASVDQLQKLITGLAEGRDPIAGAIG
MSMEG_5899|M.smegmatis_MC2_155 QLISDVIANLNTVLGTVDEKGAQFDASVDQLQQLITGLAENKDPIAGAIE
TH_1936|M.thermoresistible__bu YVINDVITHLNAVVGTLDEKSAEFDASVDQLQQLITGLAEGRDPIAGALP
MAB_4152c|M.abscessus_ATCC_199 QLIGDVINHLNEVLATVDSKSTQFSSSVDQLQQLISGLAQQRDVVAGALP
Mb3528c|M.bovis_AF2122/97 QLIGEVITNLNAVLATVDAKSAQFSASVDQLQQLVSGLAKNRDPIAGAIS
Rv3498c|M.tuberculosis_H37Rv QLIGEVITNLNAVLATVDAKSAQFSASVDQLQQLVSGLAKNRDPIAGAIS
MMAR_4986|M.marinum_M QLIGDTITNLNSVLATVDQRSAQFSASVDQLQQLITGLAENKDVIAGAIP
MUL_4061|M.ulcerans_Agy99 QLIGDTITNLNSVLATVDQRSAQFSASVDQLQQLITGLAENKDVIAGAIP
MAV_0658|M.avium_104 QLIGDVINNLNTVLTTVDQRSAQFSASVDQLQQLITGLAKHKDDIAGAIP
MLBr_02590|M.leprae_Br4923 QAIGEVVKNLNVVLDTTVRHRKEFDQTVNNLEILITGLKDHGDQLAGSFA
*.:.: :** *: * : :*. :*::*: *::** . * :**::
Mflv_1557|M.gilvum_PYR-GCK PLATAENDLTEMLETSRRPIQGVIENVRPFAQRMDERKADVNKVIEPLAE
Mvan_5201|M.vanbaalenii_PYR-1 PLATAENDLTAMLEASRRPVQGVIENVRPFAQRMDERKGDVNKVIEPLAE
MSMEG_5899|M.smegmatis_MC2_155 PLASAESDLTEMLEKSRRPVQGVIENVRPMAGRMDERKDDINKVIEPLAE
TH_1936|M.thermoresistible__bu PLASAENDLTDMLRNGRRPVQGVLENLRPLATEIDNRKEEVNRVIEPLAE
MAB_4152c|M.abscessus_ATCC_199 PLASTASSLESVLKDTRPYIKGTIEQARQLATRVDERKADVNVVVENLAE
Mb3528c|M.bovis_AF2122/97 PLASTTTDLTELLRNSRRPLQGILENARPLATELDNRKAEVNNDIEQLGE
Rv3498c|M.tuberculosis_H37Rv PLASTTTDLTELLRNSRRPLQGILENARPLATELDNRKAEVNNDIEQLGE
MMAR_4986|M.marinum_M PLASTTTDLTELLKNSRRPLQGLLENARPLATELDNRKDEVNNDVEQLGE
MUL_4061|M.ulcerans_Agy99 PLASTTTDLTELLKNSRRPLQGLLENARPLATELDNRKDEVNNDVEQLGE
MAV_0658|M.avium_104 PLASTTTDLTELLQNSRRPLQGVLENTRPLATELDNRKAEVNNDVEQLGE
MLBr_02590|M.leprae_Br4923 HISNAAGTVANLLAEDHALLHKSINYLDGIQQPLIDQRLQLEDYLHKLPT
::.: : :* : :: :: : : ::: ::: :. *
Mflv_1557|M.gilvum_PYR-GCK NYLRLN-ALGAYGSFFNIFYCSTRLKINGPAGSDILVPFGGPPDPSKGRC
Mvan_5201|M.vanbaalenii_PYR-1 NYLRLN-ALGAYGSFFNIFYCSTRLKINGPAGSDILVPFGGPPDPSKGRC
MSMEG_5899|M.smegmatis_MC2_155 NYLRLN-ALGAYGSFFNIFYCSVRLKVNGPAGSDILIPFGGPPDPSKGRC
TH_1936|M.thermoresistible__bu NYLRLQ-SLGAYGAYFNIFYCSTRIKINGPAGSDILIPFGGPPDPSKGRC
MAB_4152c|M.abscessus_ATCC_199 NYLRLN-ALGAYGSFFNIYYCSIRIKVNGPVGTDWLIPFGGPPDPSKGRC
Mb3528c|M.bovis_AF2122/97 DYLRLS-ALGSYGAFFNIYFCSVTIKINGPAGSDILLPIGGQPDPSKGRC
Rv3498c|M.tuberculosis_H37Rv DYLRLS-ALGSYGAFFNIYFCSVTIKINGPAGSDILLPIGGQPDPSKGRC
MMAR_4986|M.marinum_M DYLRLA-ALGSYGAFFNIYFCTVTIKINGPAGSDILIPMGGQPDPSKGRC
MUL_4061|M.ulcerans_Agy99 DYLRLA-ALGSYGAFFNIYFCTVTIKINGPAGSDILIPMGGQPDPSKGRC
MAV_0658|M.avium_104 DYLRLA-ALGAYGSFFNIYFCSVTIKINGPAGSDILLPLGGQVDPSKGRC
MLBr_02590|M.leprae_Br4923 VLNQIGRTIGSYGDFVNFYSCDITLKINGLQPGGPVRTVRLFQQP-TGRC
:: ::*:** :.*:: * :*:** . : .. :* .***
Mflv_1557|M.gilvum_PYR-GCK SEDG-
Mvan_5201|M.vanbaalenii_PYR-1 AEVE-
MSMEG_5899|M.smegmatis_MC2_155 SENG-
TH_1936|M.thermoresistible__bu SEDG-
MAB_4152c|M.abscessus_ATCC_199 AFKNE
Mb3528c|M.bovis_AF2122/97 AFAK-
Rv3498c|M.tuberculosis_H37Rv AFAK-
MMAR_4986|M.marinum_M AFAK-
MUL_4061|M.ulcerans_Agy99 AFAK-
MAV_0658|M.avium_104 AFVK-
MLBr_02590|M.leprae_Br4923 TPQ--
: