For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVASTPISEDLIERIVAREPRIDFIRDQSLLPPQRFAGDHAGDPAFRRTADQQRQFEQLVDSAQALYGLP DEQPAALRRTVRSNPNLLWVHTMAAGGGGQIKAAELEEDELNRITFTTSAGVHAEPLAEYALFGLLAGAK TLPRLLRQQRERKWTDRWEMGLLSQQRILLVGLGGIGRVIARKLAALGVTVVGTSRSGEPVEGVAELVHP DDLVDAVRDVDGIVVSLPGTAATENMVNAKVLRAAKPGFTLVSLGRGTVIDEPALIDALRDGQVGFAALD VFAAEPLAPESPLWSDEKVLISPHTAALNSAEDRLIADLFAENATRFLDGSPMRNRVDTVEFY
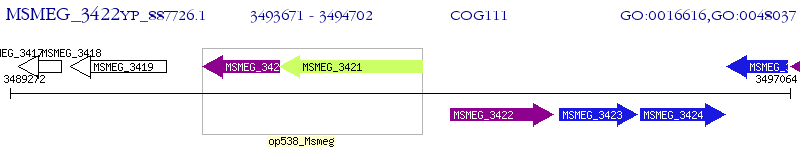
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3422 | - | - | 100% (343) | D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding |
| M. smegmatis MC2 155 | MSMEG_2496 | - | 2e-36 | 38.34% (253) | NAD-binding protein |
| M. smegmatis MC2 155 | MSMEG_0604 | - | 1e-22 | 29.39% (262) | glyoxylate reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3020c | serA1 | 4e-23 | 35.12% (205) | D-3-phosphoglycerate dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0334 | - | 7e-25 | 30.89% (259) | D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding |
| M. tuberculosis H37Rv | Rv2996c | serA1 | 4e-23 | 35.12% (205) | D-3-phosphoglycerate dehydrogenase |
| M. leprae Br4923 | MLBr_01692 | serA | 9e-22 | 35.10% (208) | D-3-phosphoglycerate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4414 | - | 3e-28 | 31.53% (314) | putative NAD-binding protein (D-isomer specific 2-hydroxyacid |
| M. marinum M | MMAR_0531 | serA3 | 5e-24 | 30.15% (262) | D-3-phosphoglycerate dehydrogenase SerA3 |
| M. avium 104 | MAV_3847 | serA | 1e-22 | 34.18% (196) | D-3-phosphoglycerate dehydrogenase |
| M. thermoresistible (build 8) | TH_2037 | serA | 1e-23 | 35.20% (196) | PROBABLE D-3-PHOSPHOGLYCERATE DEHYDROGENASE SERA1 (PGDH) |
| M. ulcerans Agy99 | MUL_1195 | serA3 | 3e-24 | 29.77% (262) | D-3-phosphoglycerate dehydrogenase SerA3 |
| M. vanbaalenii PYR-1 | Mvan_0404 | - | 2e-24 | 31.42% (261) | D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3020c|M.bovis_AF2122/97 ----MSLPVVLIAD----KLAPSTVAALGDQVEVRWVDGPDRDKLLAAVP
Rv2996c|M.tuberculosis_H37Rv ----MSLPVVLIAD----KLAPSTVAALGDQVEVRWVDGPDRDKLLAAVP
MLBr_01692|M.leprae_Br4923 ----MDLPVVLIAD----KLAQSTVAALGDQVEVRWVDGPDRTKLLAAVP
MAV_3847|M.avium_104 ----MNLPVVLIAD----KLAESTVAALGDQVEVRWVDGPDREKLLAAVP
TH_2037|M.thermoresistible__bu ----VSLPVVLIAD----KLAESTVAALGDQVEVRWVDGPDREKLLAAVA
Mflv_0334|M.gilvum_PYR-GCK ----MRVLAHFLPG----PKVTEFLSPHTDWLDIRYCAEDDDETFYRELP
Mvan_0404|M.vanbaalenii_PYR-1 ----MRVLAHFVPGGPD-SKVIEFLAPCRDWLDVRFCAEDDDDTFYRELA
MMAR_0531|M.marinum_M MNQPLRVLAHFVPG----EKVLEFVAAQRDWLDIRWCAEDDDSGFYRELD
MUL_1195|M.ulcerans_Agy99 MNQPLRVLAHFVPG----EKVLEFVAAQRDWLDIRWCAEDDDSGFYRELD
MAB_4414|M.abscessus_ATCC_1997 MNEQLVRRPVIAVQHSP-DSVPERLAPVTAAAEVRLVES---DRLATALP
MSMEG_3422|M.smegmatis_MC2_155 MVASTPISEDLIERIVAREPRIDFIRDQSLLPPQRFAGDHAGDPAFRRTA
: . : *
Mb3020c|M.bovis_AF2122/97 EADALLVRSATTVDA-------------EVLAAAPKLKIVARAGVG----
Rv2996c|M.tuberculosis_H37Rv EADALLVRSATTVDA-------------EVLAAAPKLKIVARAGVG----
MLBr_01692|M.leprae_Br4923 EADALLVRSATTVDA-------------EVLAAAPKLKIVARAGVG----
MAV_3847|M.avium_104 EADALLVRSATTVDA-------------EVLAAAPKLKIVARAGVG----
TH_2037|M.thermoresistible__bu DADALLVRSATTVDA-------------EVLAAAPKLKIVARAGVG----
Mflv_0334|M.gilvum_PYR-GCK DAEVIWH-VLRPISG-------------DDLSKAPKLRLVHKLGAG----
Mvan_0404|M.vanbaalenii_PYR-1 DAEVIWH-VLRPLSG-------------EDLARAPRLRLVHKLGAG----
MMAR_0531|M.marinum_M HAEVIWH-VLRPLSG-------------ADLQRAPRLRLVQKLGAG----
MUL_1195|M.ulcerans_Agy99 HAEVIWH-VLRPLSG-------------ADLQRAPRLRLVQKLGAG----
MAB_4414|M.abscessus_ATCC_1997 GAEVLFVYDFRSSALR------------ETWSAADSLRWVQIASTG----
MSMEG_3422|M.smegmatis_MC2_155 DQQRQFEQLVDSAQALYGLPDEQPAALRRTVRSNPNLLWVHTMAAGGGGQ
: . * * ..*
Mb3020c|M.bovis_AF2122/97 LDNVDVDAATARGVLVVNAPTSNIHSAAEHALALLLAASRQIPAADASLR
Rv2996c|M.tuberculosis_H37Rv LDNVDVDAATARGVLVVNAPTSNIHSAAEHALALLLAASRQIPAADASLR
MLBr_01692|M.leprae_Br4923 LDNVDVDAATARGVLVVNAPTSNIHSAAEHALALLLAASRQIAEADASLR
MAV_3847|M.avium_104 LDNVDVDAATERGVLVVNAPTSNIHSAAEHALALLLAAAREIPAADASLR
TH_2037|M.thermoresistible__bu LDNVDVEAATARGVLVVNAPTSNIHSAAEHAIALLLAAARQIPAADATLR
Mflv_0334|M.gilvum_PYR-GCK VNTVDVDTATARGIAVANMPGANAPSVAEGTLLLMLAALRRLPELDRATR
Mvan_0404|M.vanbaalenii_PYR-1 VNTIDVDAATARGVAVANMPGANAESVAEGTLLLMLAALRRLPVLDRATR
MMAR_0531|M.marinum_M VNTIDVATATKHGIAVANMPGANAPSVAEGTVLLMLAALRRLPELDAATR
MUL_1195|M.ulcerans_Agy99 VNTIDVATATKHGIAVANMPGANAPSVAEGTVLLMLAALRRLPELDAATR
MAB_4414|M.abscessus_ATCC_1997 VDPVLFDEFVASDVVLTNSRGIFERPIAEYVLAQILAFAKDIR---RSTR
MSMEG_3422|M.smegmatis_MC2_155 IKAAELEEDELNRITFTTSAGVHAEPLAEYALFGLLAGAKTLPRLLRQQR
:. . : ... . ** .: :** : : *
Mb3020c|M.bovis_AF2122/97 -EHTWKRSSFSGTEIFGK---TVGVVGLGRIGQLVAQRIAAFGAYVVAYD
Rv2996c|M.tuberculosis_H37Rv -EHTWKRSSFSGTEIFGK---TVGVVGLGRIGQLVAQRIAAFGAYVVAYD
MLBr_01692|M.leprae_Br4923 -AHIWKRSSFSGTEIFGK---TVGVVGLGRIGQLVAARIAAFGAHVIAYD
MAV_3847|M.avium_104 -EHTWKRSSFSGTEIFGK---TVGVVGLGRIGQLVAQRLSAFGTHLIAYD
TH_2037|M.thermoresistible__bu -QRTWKRSEFSGVEIYDK---TVGVVGLGRIGQLVAQRLAAFGTHLIAYD
Mflv_0334|M.gilvum_PYR-GCK TGNGWPSDPTLGESVRDLGGCTVGLVGYGNIAKQVERILLAIGAEVVHTS
Mvan_0404|M.vanbaalenii_PYR-1 EGRGWPSDPTLGETVRDIGGCTVGLVGYGNIAKQVERILVAMGADVIHTS
MMAR_0531|M.marinum_M QGRGWPTDPQLGESVRDIGSCTVGLIGYGNIAKRVATIVSAMGATVLHTS
MUL_1195|M.ulcerans_Agy99 QGRGWPTDPQLGESVRDIGSCTVGLIGYGNIAKRVATIVSAMGATVLHTS
MAB_4414|M.abscessus_ATCC_1997 AQAALSWRHFESESIEGT---PVGVLGTGPIGQAISTLLGAVGMRVTVLG
MSMEG_3422|M.smegmatis_MC2_155 -ERKWTDRWEMGLLSQQR----ILLVGLGGIGRVIARKLAALGVTVVGTS
. : ::* * *.: : : *.* : .
Mb3020c|M.bovis_AF2122/97 PYVSPARAAQLGIELLSLDDLLARADFISVHLPKTPETAGLIDKEALAKT
Rv2996c|M.tuberculosis_H37Rv PYVSPARAAQLGIELLSLDDLLARADFISVHLPKTPETAGLIDKEALAKT
MLBr_01692|M.leprae_Br4923 PYVAPARAAQLGIELMSFDDLLARADFISVHLPKTPETAGLIDKEALAKT
MAV_3847|M.avium_104 PYVSPARAAQLGIELLSLDELLARADFISVHLPKTPETAGLIDKDALAKT
TH_2037|M.thermoresistible__bu PYVSPARAAQLGIELVDLDELLGRADIISVHLPKTPETAGLIGADALAKV
Mflv_0334|M.gilvum_PYR-GCK TRDDGLPGWRT------LPDLLSESDIVSLHLPLTGATEGLLGRKALAQM
Mvan_0404|M.vanbaalenii_PYR-1 TRDDGSAGWRS------LPDLLSACDIVSLHLPLTDATAGLLGRDALARM
MMAR_0531|M.marinum_M TRDDGRPGWRP------LPELLATSDIVSLHLPLTPHTDRLLDADALAQM
MUL_1195|M.ulcerans_Agy99 TRDDGRPGWRP------LPELLATSDIVSLHLPLTPHTDRLLDADALARM
MAB_4414|M.abscessus_ATCC_1997 RAESAD-----------LSSHVGEFEYLVLAAPLTPQTRGIVNARVLSAM
MSMEG_3422|M.smegmatis_MC2_155 RSGEPVEGVAELVHPDDLVDAVRDVDGIVVSLPGTAATENMVNAKVLRAA
: . : : : : * * * ::. .*
Mb3020c|M.bovis_AF2122/97 KPGVIIVNAARGGLVDEAALADAITGGHVRAAGLDVFATEPCT-DSPLFE
Rv2996c|M.tuberculosis_H37Rv KPGVIIVNAARGGLVDEAALADAITGGHVRAAGLDVFATEPCT-DSPLFE
MLBr_01692|M.leprae_Br4923 KPGVIIVNAARGGLVDEVALADAVRSGHVRAAGLDVFATEPCT-DSPLFE
MAV_3847|M.avium_104 KPGVIIVNAARGGLVDEAALADAVRSGHVRAAGLDVFSKEPCT-DSPLFE
TH_2037|M.thermoresistible__bu KPGVIIVNAARGGLIDEQALADAIISGRVRAAGIDVFAKEPCT-DSPLFD
Mflv_0334|M.gilvum_PYR-GCK KTDAVLVNTSRGAIVDEAALVDALRAGTLAAAGLDVFAVEPVPSDNPLLG
Mvan_0404|M.vanbaalenii_PYR-1 KSDAVLVNTSRGPIVDEEALADALRTGKLAAAGLDVFAVEPVPADNPLLR
MMAR_0531|M.marinum_M KPNSLLINTSRGAVIDEDALVDALRSGSLAAAGLDVFAVEPVVPENPLLR
MUL_1195|M.ulcerans_Agy99 KPNSLLINTSRGAVVDEDALVDALRSGSLAAAGLDVFAVEPVVPENPLLR
MAB_4414|M.abscessus_ATCC_1997 RPTARLINVGRGELVGTWDLVSALNRGGIAGAALDVTDPEPLPVGHPLWR
MSMEG_3422|M.smegmatis_MC2_155 KPGFTLVSLGRGTVIDEPALIDALRDGQVGFAALDVFAAEPLAPESPLWS
:. ::. .** ::. * .*: * : *.:** ** **
Mb3020c|M.bovis_AF2122/97 LAQVVVTPHLGASTAEAQDRAGTDVAESVRLALAGEFVPDAVNVGGGVVN
Rv2996c|M.tuberculosis_H37Rv LAQVVVTPHLGASTAEAQDRAGTDVAESVRLALAGEFVPDAVNVGGGVVN
MLBr_01692|M.leprae_Br4923 LSQVVVTPHLGASTAEAQDRAGTDVAESVRLALAGEFVPDAVNVDGGVVN
MAV_3847|M.avium_104 LPQVVVTPHLGASTTEAQDRAGTDVAASVKLALAGEFVPDAVNVAGGTVN
TH_2037|M.thermoresistible__bu LPQVVVTPHLGASTVEAQDRAGTDVAASVKLALAGEFVPDAVNVGAGEVS
Mflv_0334|M.gilvum_PYR-GCK LPNVVLTPHVTWYTADTMRRYLEFALDNCERLRDGRALAHVVNDPSG---
Mvan_0404|M.vanbaalenii_PYR-1 LPNVVLTPHVTWYTADTMRRYLEFAVDNCERLRDGRNLVNVVNDVAG---
MMAR_0531|M.marinum_M LDNVVLTPHVTWYTVDTMRRYLTEAVDNCRRLRDGLSLANVVNCQADC--
MUL_1195|M.ulcerans_Agy99 LDNVVLTPHVTWYTVDTMRRYLTEAVDNCRRLRDGLSLANAVNCQADC--
MAB_4414|M.abscessus_ATCC_1997 TPNTHITPHNSGDVRGWSDRLQDQFVVNFERYLRGEELLNIVDKNRMHRH
MSMEG_3422|M.smegmatis_MC2_155 DEKVLISPHTAALNSAEDRLIADLFAENATRFLDGSPMRNRVDTVEFY--
:. ::** . * : . *:
Mb3020c|M.bovis_AF2122/97 EEVAPWLDLVRKLGVLAGVLSDELPVSLSVQVRGELAAEEVEVLRLSALR
Rv2996c|M.tuberculosis_H37Rv EEVAPWLDLVRKLGVLAGVLSDELPVSLSVQVRGELAAEEVEVLRLSALR
MLBr_01692|M.leprae_Br4923 EEVAPWLDLVCKLGVLVAALSDELPASLSVHVRGELASEDVEILRLSALR
MAV_3847|M.avium_104 EEVAPWLDLVRKLGLLAATLSDGPPVSLSVQVRGELASEDVEVLKLSALR
TH_2037|M.thermoresistible__bu EEVAPWVDLARKLGLLVGTLSGEPTATLQVRVAGELASEDVEVLRLSALR
Mflv_0334|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0404|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0531|M.marinum_M --------------------------------------------------
MUL_1195|M.ulcerans_Agy99 --------------------------------------------------
MAB_4414|M.abscessus_ATCC_1997 A-------------------------------------------------
MSMEG_3422|M.smegmatis_MC2_155 --------------------------------------------------
Mb3020c|M.bovis_AF2122/97 GLFSAVIEDAVTFVNAPALAAERGVTAEICKASESPNHRSVVDVRAVGAD
Rv2996c|M.tuberculosis_H37Rv GLFSAVIEDAVTFVNAPALAAERGVTAEICKASESPNHRSVVDVRAVGAD
MLBr_01692|M.leprae_Br4923 GLFSTVIEDAVTFVNAPALAAERGVSAEITTGSESPNHRSVVDVRAVASD
MAV_3847|M.avium_104 GLFSAVVEGPVTFVNAPALAEERGVTAEISTASESPNHRSVVDVRAVSHD
TH_2037|M.thermoresistible__bu GLFSTLTDQQVTYVNAPTLAAERGMQAELEKVSESPNYRSVVDVRAVSTD
Mflv_0334|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0404|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0531|M.marinum_M --------------------------------------------------
MUL_1195|M.ulcerans_Agy99 --------------------------------------------------
MAB_4414|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_3422|M.smegmatis_MC2_155 --------------------------------------------------
Mb3020c|M.bovis_AF2122/97 GSVVTVSGTLYGPQLSQKIVQINGRHFDLRAQGINLIIHYVDRPGALGKI
Rv2996c|M.tuberculosis_H37Rv GSVVTVSGTLYGPQLSQKIVQINGRHFDLRAQGINLIIHYVDRPGALGKI
MLBr_01692|M.leprae_Br4923 GSVVNIAGTLSGPQLVQKIVQVNGRNFDLRAQGMNLVIRYVDQPGALGKI
MAV_3847|M.avium_104 GTVVNVAGTLSGPQLVEKIVQINGRNFDLRARGINLVINYVDQPGALGKI
TH_2037|M.thermoresistible__bu GTVTNVAGTLSGPQQVEKIVQINGRNFDLRAEGVNLVMHYVDRPGALGKI
Mflv_0334|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0404|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0531|M.marinum_M --------------------------------------------------
MUL_1195|M.ulcerans_Agy99 --------------------------------------------------
MAB_4414|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_3422|M.smegmatis_MC2_155 --------------------------------------------------
Mb3020c|M.bovis_AF2122/97 GTLLGTAGVNIQAAQLSEDAEGPGATILLRLDQDVPDDVRTAIAAAVDAY
Rv2996c|M.tuberculosis_H37Rv GTLLGTAGVNIQAAQLSEDAEGPGATILLRLDQDVPDDVRTAIAAAVDAY
MLBr_01692|M.leprae_Br4923 GTLLGAAGVNIQAAQLSEDTEGPGATILLRLDQDVPGDVRSAIVAAVSAN
MAV_3847|M.avium_104 GTLLGSAGVNIQAAQLSEDAEGPGATILLRLDQDVPADVRSAIGAAVGAN
TH_2037|M.thermoresistible__bu GTLLGAADVNILAAQLSQDISGDGATLMLRVNRDVPADVRDAIGAAVGAS
Mflv_0334|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0404|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0531|M.marinum_M --------------------------------------------------
MUL_1195|M.ulcerans_Agy99 --------------------------------------------------
MAB_4414|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_3422|M.smegmatis_MC2_155 --------------------------------------------------
Mb3020c|M.bovis_AF2122/97 KLEVVDLS
Rv2996c|M.tuberculosis_H37Rv KLEVVDLS
MLBr_01692|M.leprae_Br4923 KLEVVNLS
MAV_3847|M.avium_104 KLEVVDLS
TH_2037|M.thermoresistible__bu TLEVVDLS
Mflv_0334|M.gilvum_PYR-GCK --------
Mvan_0404|M.vanbaalenii_PYR-1 --------
MMAR_0531|M.marinum_M --------
MUL_1195|M.ulcerans_Agy99 --------
MAB_4414|M.abscessus_ATCC_1997 --------
MSMEG_3422|M.smegmatis_MC2_155 --------