For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAPRTASKSSSPARTAKAAATRADFVRPKTAQQAVAEVLRRDITSGKLAPGSWIVQESLAEQFGLSRIPI REALKTLEAEEYITYVPHSGYRVAKLGLDELLEVFRLRDLLEAELIRDAMPAVTDDIVDLMREQMDDMDR AAEKGDLIALGLANRQFHFLTFETSTLARTRRIVTQLWNTADAYRPLYAHLMDLSKVNTEHILLVDAFVA RDAERAVAINHEHRQQAIGHLHTVFGDEKSEDAS
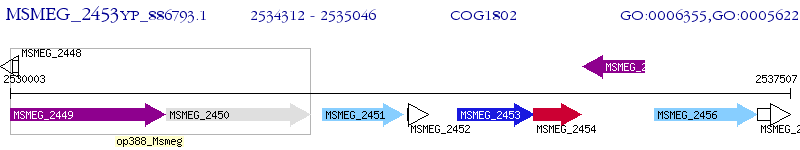
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2453 | - | - | 100% (244) | transcriptional regulator, GntR family protein |
| M. smegmatis MC2 155 | MSMEG_5731 | - | 5e-23 | 31.28% (211) | transcriptional regulator, GntR family protein |
| M. smegmatis MC2 155 | MSMEG_1317 | - | 5e-19 | 31.53% (203) | transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0170c | - | 1e-09 | 29.32% (191) | GntR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0715 | - | 1e-16 | 31.61% (155) | regulatory protein GntR, HTH |
| M. tuberculosis H37Rv | Rv0165c | - | 2e-09 | 30.65% (124) | GntR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2933c | - | 2e-19 | 29.50% (200) | GntR family transcriptional regulator |
| M. marinum M | MMAR_0408 | - | 4e-10 | 29.77% (131) | GntR family transcriptional regulator |
| M. avium 104 | MAV_0339 | - | 8e-13 | 28.06% (196) | transcriptional regulator, GntR family protein |
| M. thermoresistible (build 8) | TH_0156 | - | 1e-13 | 28.72% (188) | POSSIBLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY |
| M. ulcerans Agy99 | MUL_1058 | - | 5e-11 | 28.86% (149) | GntR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0931 | - | 6e-23 | 31.19% (202) | GntR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0170c|M.bovis_AF2122/97 MIKHDVVWVTLWPERPNNKPPPSPRQVPGNPGPTLKVLASHVNAPLSAKP
Rv0165c|M.tuberculosis_H37Rv MIKHDVVWVTLWPERPNNKPPPSPRQVPGNPGPTLKVLASHVNAPLSAKP
MMAR_0408|M.marinum_M -----------------------------------------MNIPLSARR
MUL_1058|M.ulcerans_Agy99 -----------------------------------------MNILLSARR
TH_0156|M.thermoresistible__bu -----------------------------------------MNAPITSRL
Mflv_0715|M.gilvum_PYR-GCK -----------------------------------MRAADAMNAPARAR-
MAB_2933c|M.abscessus_ATCC_199 ----------------------------------------MSRTFRYRRL
Mvan_0931|M.vanbaalenii_PYR-1 ----------------------------------------MPHR-RHSVL
MAV_0339|M.avium_104 --------------------------------------------------
MSMEG_2453|M.smegmatis_MC2_155 -------------------------------MAPRTASKSSSPARTAKAA
Mb0170c|M.bovis_AF2122/97 RSQLP-LRRAQLSDEVAGHLRAAIMSGALRSGTFIRLDETAAELGVSVTP
Rv0165c|M.tuberculosis_H37Rv RSQLP-LRRAQLSDEVAGHLRAAIMSGALRSGTFIRLDETAAELGVSVTP
MMAR_0408|M.marinum_M KSRRP-LRRAQLSDEVAGHLRAAIMSGTLRPGTFIRLDETAAELGVSITP
MUL_1058|M.ulcerans_Agy99 ESRRP-LRRAQLSDEVAGHLRAAIMSGTLRPGTFIRLDETAAELGVSITP
TH_0156|M.thermoresistible__bu GPNGGGRRRDQLSDEVAAQLRAEIMSGALRPGTYIRLDETAARLGVSITP
Mflv_0715|M.gilvum_PYR-GCK --TAGAVRREQLSDEVAARLRVEIMTGALRPESFIRLDETAARLGVSITP
MAB_2933c|M.abscessus_ATCC_199 INELRATRSGHDQTSVLDGLRRAILDGAAPPGSMIPIGEVADTYQISPIP
Mvan_0931|M.vanbaalenii_PYR-1 LGSLTVERPGGSQQVILDELRRAILDGAVPPGTPIPLAEVAEHFGVSRIP
MAV_0339|M.avium_104 --MAAIDISGTVRERAARELRDRILTGALPAGSRIDLDAITAEFATSRTP
MSMEG_2453|M.smegmatis_MC2_155 ATRADFVRPKTAQQAVAEVLRRDITSGKLAPGSWIVQESLAEQFGLSRIP
** * * . : * : * *
Mb0170c|M.bovis_AF2122/97 VREALLKLRGEGMVGLEPHRGHVVLPLTRQDIDDIFWLQATIAQELATSA
Rv0165c|M.tuberculosis_H37Rv VREALLKLRGEGMVGLEPHRGHVVLPLTRQDIDDIFWLQATIAQELATSA
MMAR_0408|M.marinum_M VREALLKLRGEGMVQLEPHRGHVVLPLTRQDIDDIFWLQATIARELAVTA
MUL_1058|M.ulcerans_Agy99 VREALLKLRGEGMVQLEPHRGHVVLPLTRQDIDDIFWLQATIARELAVTA
TH_0156|M.thermoresistible__bu IREALRTLRGEGMVQLEPHRGHVVAPLTRGDIEDIFWLQATIARKLTAAA
Mflv_0715|M.gilvum_PYR-GCK VREALRTLRGEGMVELEPHRGHRVIPLSRNDIEDIFWLQSTIAQELAATA
MAB_2933c|M.abscessus_ATCC_199 VREALKTLVAEGLVDHRANGGYRVAQLTLPELREIYFTRGVLERAALAQA
Mvan_0931|M.vanbaalenii_PYR-1 VRESLKTLIGEGLVAHRANSGYMVAQLTAAELGELYVARESLEAASLAAA
MAV_0339|M.avium_104 VREALLELSFEGLVQVAPRSGVTVIGISPEDVLDSFTILGVLTGQAAAWA
MSMEG_2453|M.smegmatis_MC2_155 IREALKTLEAEEYITYVPHSGYRVAKLGLDELLEVFRLRDLLEAELIRDA
:**:* * * : .. * * : :: : : : *
Mb0170c|M.bovis_AF2122/97 TAHITDVEIDELDRINNALAGAIGSGDAKTIASIEFAFHRVFNKASRRIK
Rv0165c|M.tuberculosis_H37Rv TAHITDVEIDELDRINNALAGAIGSGDAKTIASIEFAFHRVFNKASRRIK
MMAR_0408|M.marinum_M TERISEIDIGELEHINDALTAAVGAGDTEAIAALEFAFHRVLNQSSGRIK
MUL_1058|M.ulcerans_Agy99 TERISEVVIGELEHINDALTAAVGAGDTEAIAALEFAFHRVLNQSSGRIK
TH_0156|M.thermoresistible__bu VDRITDAELDELTALNEALAAAVDRGVVGEVAAAMSAFHRVLHRTTGRIK
Mflv_0715|M.gilvum_PYR-GCK TRRITEEQIEQLSRLNDTLASAVEQHDVQRIAQAEFEFHRTFNRAAGRVK
MAB_2933c|M.abscessus_ATCC_199 AARATPEDVDRALAEHQLLQQATVEDDARAFHVHSRRFHRALVLPCGMHR
Mvan_0931|M.vanbaalenii_PYR-1 VGNASEADFATAIDMNALLEHAIREDDAVAYHRQSRRFHAALTGPSRMYR
MAV_0339|M.avium_104 AERIGPEELATLRELAAEVVAKSGDD---SIGEANWHFHQMIHRAAHSPR
MSMEG_2453|M.smegmatis_MC2_155 MPAVTDDIVDLMREQMDDMDRAAEKGDLIALGLANRQFHFLTFETSTLAR
. : ** . :
Mb0170c|M.bovis_AF2122/97 LAWFLLNAARYMP--AQVFAADPRWGADAVNSHRQLIAALRRRDTAAVIE
Rv0165c|M.tuberculosis_H37Rv LAWFLLNAARYMG--AGVRGRP----AMGRGRGEQSSAADRR---AAPPR
MMAR_0408|M.marinum_M LAWFLLNAARYMP--AQLYASDPQWGAAAVDNHRQLIAALRRRDTAAVIE
MUL_1058|M.ulcerans_Agy99 LAWFLLNAARYMP--AQLYASDPQWGAAAVDNHRQLIAALRRRDTAAVIE
TH_0156|M.thermoresistible__bu LAWFLYHVARYMP--PQIYATDQQWGTATVARHRELIAALRRRDTETATR
Mflv_0715|M.gilvum_PYR-GCK LSWFLLHAARYLP--PQLFASDDDWGAQAVVSHAELIAALRRGDVDVVVE
MAB_2933c|M.abscessus_ATCC_199 LLHMFEATWNLTEPFQVMRTVSASDQLQLHADHEAMITAAASRDIDTLLS
Mvan_0931|M.vanbaalenii_PYR-1 LLRMLETAWNLTEPVQPMVHVTPDDRIRLHADHGEMLQAFLTRDVERLLG
MAV_0339|M.avium_104 LANQIKHAARVVP--TNFLTLFPEHEKHSLDEHAQLLDALGDKDVERARA
MSMEG_2453|M.smegmatis_MC2_155 TRRIVTQLWNTADAYRPLYAHLMDLSKVNT-EHILLVDAFVARDAERAVA
. . . *
Mb0170c|M.bovis_AF2122/97 HTVWQFTDGARRLTEALAETEVFG---------
Rv0165c|M.tuberculosis_H37Rv HSRR---NRAHRLAVHRWGTQADGGPG------
MMAR_0408|M.marinum_M HSVWQFTDAAHRLTQMLETNGIFN---------
MUL_1058|M.ulcerans_Agy99 HSVWQFTDAAHRLTQMLETNGIFN---------
TH_0156|M.thermoresistible__bu VTVDLFTDSAQRLIAQLEKVGIWT---------
Mflv_0715|M.gilvum_PYR-GCK LTRGQFTDGLHRLIAWLEQVGLWS---------
MAB_2933c|M.abscessus_ATCC_199 VAATHHDRLESVIVAAAGQLDVVAEA-------
Mvan_0931|M.vanbaalenii_PYR-1 AAERHARRLNSVIATLSFETGLLAAEKDISSPQ
MAV_0339|M.avium_104 IAERHVLDAGRSLAEWLEQRRDGER--------
MSMEG_2453|M.smegmatis_MC2_155 INHEHRQQAIGHLHTVFGDEKSEDAS-------
: