For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ASPATGGGRPRRERGSITVDEILSGAFEVAREVSVDNLSMPQLAKHLDVGVTSIYWYFRRKDDLLDAMTE LALREYDFGVSSIDAGSWRESLRAHAHRMRDTIQQNPILCDLILIRGTGGTPAARHALEKIEQPVAALVQ AGLSPQQAVQTYSAIQVLVRSSAVLHRLQNRASTVPLPRDHWQQVIDPQTMPLISSLTGDGYRIGAADTA NFDHILDSILERAEHLTASPRDSSPRADTILRENH*
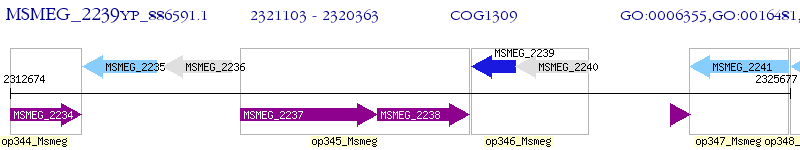
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2239 | - | - | 100% (246) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_2225 | - | 2e-60 | 54.55% (220) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_2583 | - | 1e-08 | 27.75% (209) | putative transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1388c | - | 1e-59 | 53.33% (225) | transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_4355 | - | 8e-59 | 51.53% (229) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv1353c | - | 1e-59 | 53.33% (225) | transcriptional regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1508c | - | 2e-46 | 40.09% (227) | putative HTH-type transcriptional regulator |
| M. marinum M | MMAR_4019 | - | 3e-61 | 52.84% (229) | transcriptional regulatory protein |
| M. avium 104 | MAV_1586 | - | 2e-61 | 54.59% (229) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_1116 | - | 4e-60 | 51.54% (227) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN |
| M. ulcerans Agy99 | MUL_3884 | - | 4e-61 | 52.40% (229) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_1989 | - | 3e-58 | 48.78% (246) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4019|M.marinum_M ---------MEVPVVAKRAAADKRQRRERGSINPDDIISGAFELAEQVSI
MUL_3884|M.ulcerans_Agy99 ---------MEVPVVAKRAAADKRQRRERGSINPDDIISGAFELAEQVSI
MAV_1586|M.avium_104 --------------MAKQATADKRQRRERGSINPDDIITGAFELAEQVSI
Mb1388c|M.bovis_AF2122/97 ----------------MQTTPGKRQRRQRGSINPEDIISGAFELAQQVSI
Rv1353c|M.tuberculosis_H37Rv ----------------MQTTPGKRQRRQRGSINPEDIISGAFELAQQVSI
Mflv_4355|M.gilvum_PYR-GCK --------------MARQATAEKRQRRERGSINPDDIIKGAFELAEQVGI
Mvan_1989|M.vanbaalenii_PYR-1 --------------MAKQATAEKRQRRERGSINPDDIIKGAFELAEQVGI
TH_1116|M.thermoresistible__bu --------------VAKQATADKRRRRERGSINPDDIVAGAFELAEEVGI
MSMEG_2239|M.smegmatis_MC2_155 --------------MASPATGGGRPRRERGSITVDEILSGAFEVAREVSV
MAB_1508c|M.abscessus_ATCC_199 MRHAGVGHEARTTSTQGPSHSPKRERRRRGSITAEDIIAGAFELAEEIGI
: * **.****. ::*: ****:*.::.:
MMAR_4019|M.marinum_M DNLSMPMLGKHLGVGVTSIYWYFRKKDDLLNAMTDRALSKYVFATPYVEA
MUL_3884|M.ulcerans_Agy99 DNLSMPMLGKHLGVGVTSIYWYFRKKDDLLNAMTDRALSKYVFATPYVEG
MAV_1586|M.avium_104 DNLSMPLLGKHLGVGVTSIYWYFRKKDDLLNAMTDRALSKYVFATPYVEA
Mb1388c|M.bovis_AF2122/97 DNLSMPLLGKHLGVGVTSIYWYFRKKDDLLNAMTDRALSKYVFATPYIEA
Rv1353c|M.tuberculosis_H37Rv DNLSMPLLGKHLGVGVTSIYWYFRKKDDLLNAMTDRALSKYVFATPYIEA
Mflv_4355|M.gilvum_PYR-GCK DNLSMPLLGKHLGVGVTSIYWYFRKKDDLLNAMTDRALRQYAFATPYVEA
Mvan_1989|M.vanbaalenii_PYR-1 DNLSMPLLGKHLGVGVTSIYWYFRKKDDLLNAMTDRALRQYAFATPYVEA
TH_1116|M.thermoresistible__bu DNLSMPLLGKHLGVGVTSIYWYFRKKDDLLNEMTNRALRQYVFETPYVEA
MSMEG_2239|M.smegmatis_MC2_155 DNLSMPQLAKHLDVGVTSIYWYFRRKDDLLDAMTELALREYDFGVSSIDA
MAB_1508c|M.abscessus_ATCC_199 SGLSMPLLAKHLDVGVTSIYWYFRKKGELLDAMTDRATKQYHFAAPFVGD
..**** *.***.***********:*.:**: **: * :* * .. :
MMAR_4019|M.marinum_M SDWRETLRNHARLMRKTFMGNPILCDLILIR-SALSPKAARLGAQEMERA
MUL_3884|M.ulcerans_Agy99 SDWRETLRNHARLMRKTFMSNPILCDLILIR-SALSPKTARLGAQKMERA
MAV_1586|M.avium_104 SDWRETLRNHARLMRKTFMGNPILCDLILIR-AALSPKAARLGAQEMEKA
Mb1388c|M.bovis_AF2122/97 GDWRETLRNHARSMRKTFADNPVLCDLILIR-AALSPKTARLGAQEMEKA
Rv1353c|M.tuberculosis_H37Rv GDWRETLRNHARSMRKTFADNPVLCDLILIR-AALSPKTARLGAQEMEKA
Mflv_4355|M.gilvum_PYR-GCK RDWRETLANHARTMRKTFNGNPILCDLILIR-SALSPRAAKVGVQQVEKA
Mvan_1989|M.vanbaalenii_PYR-1 KDWRETLANHARSMRQTFNGNPILCDLILIR-SALSPRAAKLGVQEVERA
TH_1116|M.thermoresistible__bu GNWRESLRNHARNMRKTFLSSPILTDLVLIR-GALNTKIRKLGLQQMEAA
MSMEG_2239|M.smegmatis_MC2_155 GSWRESLRAHAHRMRDTIQQNPILCDLILIR-GTGGTPAARHALEKIEQP
MAB_1508c|M.abscessus_ATCC_199 ETWQDALRSHTHRMREAFRERPVLVELILMRTSELSSEALQASIQNLESL
*:::* *:: **.:: *:* :*:*:* . .. : . :::*
MMAR_4019|M.marinum_M IANLVEAGLSPEDAFDTYAAVSIHVRGSVVLHRLYEKTQSADSGPRAIED
MUL_3884|M.ulcerans_Agy99 IANLVEAGLSPEDAFDTYAAVSIHVRGSVVLHRLYEKTQSADSGPRAIED
MAV_1586|M.avium_104 VANLVQAGLSPEDAFDTYSAISVHVRGSVVLQRLYEKNQSSDVGPRAIED
Mb1388c|M.bovis_AF2122/97 IANLVTAGLSLEDAFDIYSAVSVHVRGSVVLDRLSRKSQSAGSGPSAIEH
Rv1353c|M.tuberculosis_H37Rv IANLVTAGLSLEDAFDIYSAVSVHVRGSVVLDRLSRKSQSAGSGPSAIEH
Mflv_4355|M.gilvum_PYR-GCK IGSLVEAGLSPEDAFDTYSAMSVHVRGSVVLQRLREKNLAGDDGHGDIDD
Mvan_1989|M.vanbaalenii_PYR-1 IGSLVEAGLSPEDAFDTYSAMSVHVRGSVVLQRLREKNLAGEDGHGDIDD
TH_1116|M.thermoresistible__bu IDSLVQAGLSPEDAFDTYSAVSVHVRGSVVLERLHEKTQDLFNGG---DE
MSMEG_2239|M.smegmatis_MC2_155 VAALVQAGLSPQQAVQTYSAIQVLVRSSAVLHRLQNR--ASTVPLPRDHW
MAB_1508c|M.abscessus_ATCC_199 VRTLIGVGFSPDNALDVYFALSLHIRGVVVLEHMDTVMPASGEAR-----
: *: .*:* ::*.: * *:.: :*. .**.::
MMAR_4019|M.marinum_M AMNIDPLTTPLLAQVSGKGHRIATPDEANFEYGLDCILDHAGRLIDERAK
MUL_3884|M.ulcerans_Agy99 AMNIDPLTTPLLAQVSGKGHRIATPDEANFEYGLDCILDHAGRLIDERAK
MAV_1586|M.avium_104 AVAIDPEKTPLLAQVTRIGHRIGAPDETNFEYGLTCILDHASRLIEE-GK
Mb1388c|M.bovis_AF2122/97 PVAIDPATTPLLAHATGRGHRIGAPDETNFEYGLECILDHAGRLIEQSSK
Rv1353c|M.tuberculosis_H37Rv PVAIDPATTPLLAHATGRGHRIGAPDETNFEYGLECILDHAGRLIEQSSK
Mflv_4355|M.gilvum_PYR-GCK TMAIDPETAPLLAAVTQKGHHLSASDDRNFEFGLDCILENAARLIEK-AK
Mvan_1989|M.vanbaalenii_PYR-1 TMTIDPESAPLLAAVTQKGHHLSASDDRNFEYGLECILDHAARLIEQNAK
TH_1116|M.thermoresistible__bu PSAVDPASTPIIAELLAKGHRLAEADERNFEYTLECILDHAERLIEERAA
MSMEG_2239|M.smegmatis_MC2_155 QQVIDPQTMPLISSLTGDGYRIGAADTANFDHILDSILERAEHLTASPRD
MAB_1508c|M.abscessus_ATCC_199 SLVPSATLAPTLHELASRGHSIATISDANFDFTVEAIIERAENLLAQDIA
.. * : *: :. . **:. : .*::.* .* .
MMAR_4019|M.marinum_M AAKSAPSR-PRKAAKPPAPRTRAKATTR-
MUL_3884|M.ulcerans_Agy99 AAKSAPSR-PRKAAKPPAPRTRAKATTR-
MAV_1586|M.avium_104 GAKAGPSR-QRKPAKSSAARGRTKAPANR
Mb1388c|M.bovis_AF2122/97 AAGEVAVRRPTATADAPTPGARAKAVAR-
Rv1353c|M.tuberculosis_H37Rv AAGEVAVRRPTATADAPTPGARAKAVAR-
Mflv_4355|M.gilvum_PYR-GCK TTRKAP------------ARKR-------
Mvan_1989|M.vanbaalenii_PYR-1 PARKATPA-------KSTARKR-------
TH_1116|M.thermoresistible__bu GGTRK------------------------
MSMEG_2239|M.smegmatis_MC2_155 SSPRADT----------ILRENH------
MAB_1508c|M.abscessus_ATCC_199 AREAS------------------------