For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ARRSVTSFDVAKKAGVSQPTVSRALRNLPGTSPETRERVRTAALELDYIPSASGRTLSTRTTQRVAIVAE ALTNPFYPELVEPLRNALGEYGYRAVLVADRADDPVTIDALADGSYDGVLLCTVTRRSTLPRDLTERGVP HVLVNRILDVPESSSCSFDNLAGGRLVGEFLAGLGHQRIGALHGTVDNSTARDRASGLRSSLRSHGLHVR RTDLRRVPFSYDAGHRAALEFLDRPDRPSAVFCGNDVIAVGALSAAKALRIRVPEELTIIGFDNIGAAGW GTVDLTTVHCDRDELARVSVELLLRAIDGAEPEQHVIAPGGMVERGTHGAPGC*
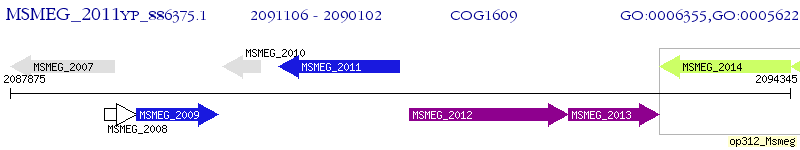
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2011 | - | - | 100% (334) | transcriptional regulator, LacI family protein |
| M. smegmatis MC2 155 | MSMEG_2989 | - | 3e-36 | 32.23% (332) | LacI family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_6376 | - | 1e-32 | 29.90% (311) | transcriptional regulator LacI family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3606c | - | 3e-16 | 28.09% (324) | transcriptional regulatory protein LacI-family |
| M. gilvum PYR-GCK | Mflv_1457 | - | 4e-17 | 29.46% (336) | periplasmic binding protein/LacI transcriptional regulator |
| M. tuberculosis H37Rv | Rv3575c | - | 3e-16 | 28.09% (324) | transcriptional regulatory protein LacI-family |
| M. leprae Br4923 | MLBr_00398 | - | 6e-06 | 25.13% (191) | putative D-ribose-binding protein |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_5070 | - | 2e-16 | 27.27% (330) | PurR family transcriptional regulator |
| M. avium 104 | MAV_0585 | - | 2e-18 | 29.54% (325) | transcriptional regulator, LacI family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4146 | - | 7e-16 | 26.97% (330) | PurR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0385 | - | 2e-33 | 29.88% (338) | periplasmic binding protein/LacI transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3606c|M.bovis_AF2122/97 --MSPTPRRRATLASLAAELKVSRTTVSNAFNRPDQLSADLRERVLATAK
Rv3575c|M.tuberculosis_H37Rv --MSPTPRRRATLASLAAELKVSRTTVSNAFNRPDQLSADLRERVLATAK
MMAR_5070|M.marinum_M --MSPTPRRRATLASLAAELKVSRSTISNAFNRPDQLSAELRERVLATAK
MUL_4146|M.ulcerans_Agy99 --MSPTPRRRATLASLAAELKVSRSTISNAFNRPDQLSVELRERVLATAK
MAV_0585|M.avium_104 --MSPTPRRRATLASLADELKVSRTTVSNAFNRPDQLSADLRERVLATAK
Mflv_1457|M.gilvum_PYR-GCK MSRSPTPRRRATLASLAAELKVSRTTVSNAYNRPDQLSAELRERVLSTAK
Mvan_0385|M.vanbaalenii_PYR-1 --MSTPRRRRPTMADVAERAGVSRTLVSFILDGKPGASDETRRRVLAIAD
MSMEG_2011|M.smegmatis_MC2_155 -----MARRSVTSFDVAKKAGVSQPTVSRALRNLPGTSPETRERVRTAAL
MLBr_00398|M.leprae_Br4923 -------------MSTGTSVPLRVASIMLILGLLPAILPACGFSESHHKL
. . : . :
Mb3606c|M.bovis_AF2122/97 RLGYAGPDPVARSLRTRKAGAVGLVMAEPLTYFFSDPAARDFVAGVAQSC
Rv3575c|M.tuberculosis_H37Rv RLGYAGPDPVARSLRTRKAGAVGLVMAEPLTYFFSDPAARDFVAGVAQSC
MMAR_5070|M.marinum_M RMGYPGPDPVARSLRTRKAGAVGLVMAEPLTYFFSDPAARDFVTGVAQSC
MUL_4146|M.ulcerans_Agy99 RMGYPGPDPVARSLRTRKAGAVGLVMAEPLTYFFSDPAARDFVTGVAQSC
MAV_0585|M.avium_104 RLGYPGPDPVARSLRTRKAGAVGLVMAEPLTYFFSDPAARDFVTGVAQSC
Mflv_1457|M.gilvum_PYR-GCK RLGYPGPDPVARSLRTRRAGAVGLVITEPLNYSFSDPAALDFVAGLAESC
Mvan_0385|M.vanbaalenii_PYR-1 EIGYR-PDSAARLLAQGRSRTLGVMTDVRQLFQA------ELVTHIYPAA
MSMEG_2011|M.smegmatis_MC2_155 ELDYI-PSASGRTLSTRTTQRVAIVAEALTNPFYP-----ELVEPLRNAL
MLBr_00398|M.leprae_Br4923 TIGVSYPTANSPFWNAYTR----------------------FIDEGSHQL
:. * . . ::
Mb3606c|M.bovis_AF2122/97 EELGQGLQLVSVGSSRSLADGTAAVLGAGVDGFVVYSVGD--DDPYLQVV
Rv3575c|M.tuberculosis_H37Rv EELGQGLQLVSVGSSRSLADGTAAVLGAGVDGFVVYSVGD--DDPYLQVV
MMAR_5070|M.marinum_M EELGQGLLLVAVGASRSLEDGTAAVLNAGVDGFVVYSVRD--DDPYLQVV
MUL_4146|M.ulcerans_Agy99 EELGQGLLLVAVGASRSLEDGTAAVLNAGVDGFVVYSVRD--DDPYLQVV
MAV_0585|M.avium_104 EALGQGLLLVAVGPSRSLDEGTAAVLSAGVDGFVVYSVCD--DDPYLQAV
Mflv_1457|M.gilvum_PYR-GCK EAVGQGLLLVAVGPNRSVAEGSAAVLAAGVDGFVVYSASD--DDPYLPAV
Mvan_0385|M.vanbaalenii_PYR-1 ERLGYEVLLSANLPDRTEATVVDSLLSHRCGALILLGPTG--DQSFLEKL
MSMEG_2011|M.smegmatis_MC2_155 GEYGYRAVLVADRADDPVT--IDALADGSYDGVLLCTVTR--RSTLPRDL
MLBr_00398|M.leprae_Br4923 GVYIN--AVSAEEDEQKQLSDVETLINQGIDGLIVTPQSTSIAPTLLRIA
: : . :: ...:: .
Mb3606c|M.bovis_AF2122/97 LQRRLPVVVVDQPKDLSGVS--------RVGIDDRAAMRELAGYVLGLGH
Rv3575c|M.tuberculosis_H37Rv LQRRLPVVVVDQPKDLSGVS--------RVGIDDRAAMRELAGYVLGLGH
MMAR_5070|M.marinum_M LQRRLPVVVVDQPKELAGVS--------RVGIDDRAAMRKIADYVLGLGH
MUL_4146|M.ulcerans_Agy99 LQRRLPVVVVDQPKELAGVS--------RVGIDDRAAMRKIADYVLSLGH
MAV_0585|M.avium_104 LQRRLPVVVVDQPKGLTGVS--------RVGIDDRAAMRELADYVLGLGH
Mflv_1457|M.gilvum_PYR-GCK MERHLPVVVVDQPRDVPGAS--------QVRIDDRAAMRSLAEHVIDLGH
Mvan_0385|M.vanbaalenii_PYR-1 AQRAPVVVACRRLTGLNQQTP-----LATVRTNDAKGIRQAVDYLVGLGH
MSMEG_2011|M.smegmatis_MC2_155 TERGVPHVLVNRILDVPESS--------SCSFDNLAGGRLVGEFLAGLGH
MLBr_00398|M.leprae_Br4923 TQANIPVVVVDRYPGYAPGQNKNADYVAFLGPNNEKAGSGIAEALMAGGG
: * : :: . : *
Mb3606c|M.bovis_AF2122/97 RELGLLTMRLGRDRRQD----LVDAERLRSPTFDVQRERIVGVWEAMTAA
Rv3575c|M.tuberculosis_H37Rv RELGLLTMRLGRDRRQD----LVDAERLRSPTFDVQRERIVGVWEAMTAA
MMAR_5070|M.marinum_M RQIGLLTMRLGRDRRQD----LVDAERLKSPTFDVQRERITGVWEAMTAA
MUL_4146|M.ulcerans_Agy99 RQIGLLTMRLGRDRRQD----LVDAERLKSPTFDVQRERITGVWEAMTAA
MAV_0585|M.avium_104 REIGLLTMRLGRDRRQG----LVDAERLRSPAFDVQRERITGVWEAMTAA
Mflv_1457|M.gilvum_PYR-GCK REIGLLTMRLGRDWPHGPGPALADPERVRAPNFHVQCERIHGVYDAMTAA
Mvan_0385|M.vanbaalenii_PYR-1 RRIHHVDG--GRD--------------------PGSADRSRAYKAAMRAH
MSMEG_2011|M.smegmatis_MC2_155 QRIGALHGTVDNS---------------------TARDRASGLRSSLRSH
MLBr_00398|M.leprae_Br4923 SKFLALGGMPGNS---------------------VAQGRKAGLESALTAV
.: : ... * . :: :
Mb3606c|M.bovis_AF2122/97 GVDPDSLTVVESYEHLPTSGGTAAKVALQANPR--LTALMCTADILALSA
Rv3575c|M.tuberculosis_H37Rv GVDPDSLTVVESYEHLPTSGGTAAKVALQANPR--LTALMCTADILALSA
MMAR_5070|M.marinum_M GVDPESLTVVESYEHLPTSGGDAAKVALEANPR--ITALMCTADILALSA
MUL_4146|M.ulcerans_Agy99 GVDPESLTVVESYEHLPTSGGDAAKVALEANPR--ITALMCTADILALSA
MAV_0585|M.avium_104 GVDPDSLTVVESYEHLPESGGAAAKVALEANPR--ITALMCTADILALSA
Mflv_1457|M.gilvum_PYR-GCK GLDPASLTLVESYEHLPTSGGAGADIALEANPR--ITALMCTADVLALSA
Mvan_0385|M.vanbaalenii_PYR-1 GLAEEIEVIPG--AHNEDAGAAAARAMLAAPTL--PTAVLAGNDRCALGI
MSMEG_2011|M.smegmatis_MC2_155 GLHVR-RTDLRRVPFSYDAGHRAALEFLDRPDR--PSAVFCGNDVIAVGA
MLBr_00398|M.leprae_Br4923 G---HRLVQFQYAGDSEDKGLAAAENMLQAHPAGDANAIWCFNDKLCQGA
* . * .* * .*: . * . .
Mb3606c|M.bovis_AF2122/97 MDYLRAHGIYVPGQMTVTGFDG--VPEALSRGLTTVAQPSLHKGHRAGEL
Rv3575c|M.tuberculosis_H37Rv MDYLRAHGIYVPGQMTVTGFDG--VPEALSRGLTTVAQPSLHKGHRAGEL
MMAR_5070|M.marinum_M MDYLRGHGIYVPGQMTVTGFDG--VPEAISRGLTTVSQPSLQKGRRAGEL
MUL_4146|M.ulcerans_Agy99 MDYLRGHGIYVPGQMTVTGFDG--VPEAISRGLTTVSQPSLQKGRRAGEL
MAV_0585|M.avium_104 MDYLRAHGVYVPGQLTVTGFDG--VPDAISRGLTTVSQSSLHKGRRAGEL
Mflv_1457|M.gilvum_PYR-GCK MDHLRAKGIYVPGQMTVTGFDG--VPEARQRGLTTVAQPGVEKGRRAGEL
Mvan_0385|M.vanbaalenii_PYR-1 LDVFTRAGVDVPAQVSLMGYDDTRLSENPRIDLSTIHQDAAEIARHAVQL
MSMEG_2011|M.smegmatis_MC2_155 LSAAKALRIRVPEELTIIGFDNIGAAGWGTVDLTTVHCDRDELARVSVEL
MLBr_00398|M.leprae_Br4923 IKAVYNANREKEFIFGGMDLTPQAIAAIENGLYTVSLGAHWLEGGFGLAI
:. . . . :. . . :
Mb3606c|M.bovis_AF2122/97 LLKPPRSGLP---VIEVLDTELVRGRTAGPPA------------------
Rv3575c|M.tuberculosis_H37Rv LLKPPRSGLP---VIEVLDTELVRGRTAGPPA------------------
MMAR_5070|M.marinum_M LQNPPRSGLP---VVELLDTELIRGRTAGPPA------------------
MUL_4146|M.ulcerans_Agy99 LQNPPRSGLP---VVELLDTELIRGRTAGPPA------------------
MAV_0585|M.avium_104 LLKPARSGLP---VVELLDTELIRGRTAGPPA------------------
Mflv_1457|M.gilvum_PYR-GCK LHNPPRSGLP---VVEVLDTEVIRGRTSGPPA------------------
Mvan_0385|M.vanbaalenii_PYR-1 AVQMLVEGPPHAMDVVLEPTLVVRGTTAPPRGGPAARSGIADTIEG----
MSMEG_2011|M.smegmatis_MC2_155 LLRAIDGAEPEQ-HVIAPGGMVERGTHGAPGC------------------
MLBr_00398|M.leprae_Br4923 LYRKIHGQDPAERMVKLDLLKVDKSNVAKFKARYIDNTPHYNWKSTTPFD
. * : : :. .
Mb3606c|M.bovis_AF2122/97 ------
Rv3575c|M.tuberculosis_H37Rv ------
MMAR_5070|M.marinum_M ------
MUL_4146|M.ulcerans_Agy99 ------
MAV_0585|M.avium_104 ------
Mflv_1457|M.gilvum_PYR-GCK ------
Mvan_0385|M.vanbaalenii_PYR-1 ------
MSMEG_2011|M.smegmatis_MC2_155 ------
MLBr_00398|M.leprae_Br4923 MTITLN