For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTQHSKSDQARKSFIDSVKGTAKEVVGAVTGNDSLTAEGQLEKTQAKERREAGRAAAVADAEARQARAEE AQAQADGAQKRAAVAAEAAVVENDVRAQQDAQKREAEAAARRDAERQKTQADIDAQREVQLADAAERSDV AVARQEAAEAAADHQDAVKESADVRARADRMRQEADKLTEEADLPGDEK
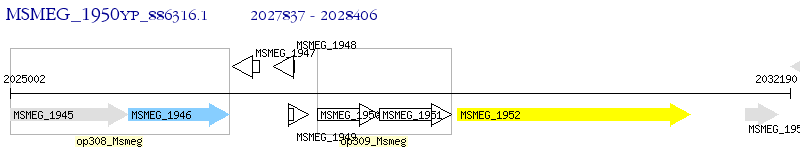
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1950 | - | - | 100% (189) | hypothetical protein MSMEG_1950 |
| M. smegmatis MC2 155 | MSMEG_1517 | - | 1e-07 | 27.32% (183) | SPFH domain-containing protein/band 7 family protein |
| M. smegmatis MC2 155 | MSMEG_1770 | - | 5e-07 | 36.71% (79) | hypothetical protein MSMEG_1770 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_5236 | - | 6e-45 | 52.51% (179) | CsbD family protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_01120 | - | 1e-06 | 30.26% (152) | hypothetical protein MLBr_01120 |
| M. abscessus ATCC 19977 | MAB_2518 | - | 1e-08 | 34.25% (73) | hypothetical protein MAB_2518 |
| M. marinum M | MMAR_2989 | - | 2e-08 | 36.84% (76) | hypothetical protein MMAR_2989 |
| M. avium 104 | MAV_4077 | - | 3e-10 | 40.51% (79) | hypothetical protein MAV_4077 |
| M. thermoresistible (build 8) | TH_4168 | - | 7e-09 | 39.51% (81) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_2033 | smc | 3e-08 | 27.93% (179) | chromosome partition protein Smc |
| M. vanbaalenii PYR-1 | Mvan_1034 | - | 4e-41 | 48.89% (180) | CsbD-like protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5236|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1034|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1950|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_2989|M.marinum_M --------------------------------------------------
MAV_4077|M.avium_104 --------------------------------------------------
TH_4168|M.thermoresistible__bu --------------------------------------------------
MAB_2518|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_2033|M.ulcerans_Agy99 MYLKSLTLKGFKSFAAPTTLRFEPGITAVVGPNGSGKSNVVDALAWVMGE
MLBr_01120|M.leprae_Br4923 --------------------------------------------------
Mflv_5236|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1034|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1950|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_2989|M.marinum_M --------------------------------------------------
MAV_4077|M.avium_104 --------------------------------------------------
TH_4168|M.thermoresistible__bu --------------------------------------------------
MAB_2518|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_2033|M.ulcerans_Agy99 QGAKTLRGGKMEDVIFAGTSLRAPLGRAEVTVTIDNSDNALPIEYTEVSI
MLBr_01120|M.leprae_Br4923 --------------------------------------------------
Mflv_5236|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1034|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1950|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_2989|M.marinum_M --------------------------------------------------
MAV_4077|M.avium_104 --------------------------------------------------
TH_4168|M.thermoresistible__bu --------------------------------------------------
MAB_2518|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_2033|M.ulcerans_Agy99 TRRMFRDGASEYEINGSSCRLMDVQELLSDSGIGREMHVIVGQGKLDEIL
MLBr_01120|M.leprae_Br4923 --------------------------------------------------
Mflv_5236|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1034|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1950|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_2989|M.marinum_M --------------------------------------------------
MAV_4077|M.avium_104 --------------------------------------------------
TH_4168|M.thermoresistible__bu --------------------------------------------------
MAB_2518|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_2033|M.ulcerans_Agy99 QSRPEDRRAFIEEAAGVLKHRKRKEKALRKLDAMSANLARLTDLTTELRR
MLBr_01120|M.leprae_Br4923 ---------------------------------MKLHRLALTNYRGTARR
Mflv_5236|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1034|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1950|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_2989|M.marinum_M --------------------------------------------------
MAV_4077|M.avium_104 --------------------------------------------------
TH_4168|M.thermoresistible__bu --------------------------------------------------
MAB_2518|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_2033|M.ulcerans_Agy99 QLKPLGRQAEVARRAQTIQADLRDARLRLAADDVVSRRAEREAIFEAEAT
MLBr_01120|M.leprae_Br4923 EIEFPDRGVILVCGANEIGKSS------------MIEALDLLLEFRDRST
Mflv_5236|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1034|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1950|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_2989|M.marinum_M --------------------------------------------------
MAV_4077|M.avium_104 --------------------------------------------------
TH_4168|M.thermoresistible__bu --------------------------------------------------
MAB_2518|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_2033|M.ulcerans_Agy99 MRREHDEAAARLTVAAEELAAHEATLAELTSRAESVQHTWFGLSALAERV
MLBr_01120|M.leprae_Br4923 KKEVKQVKPANADIGSEVCAEISSGTYRFVYRKRFNKKCETALTVLAPHR
Mflv_5236|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1034|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1950|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_2989|M.marinum_M --------------------------------------------------
MAV_4077|M.avium_104 --------------------------------------------------
TH_4168|M.thermoresistible__bu --------------------------------------------------
MAB_2518|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_2033|M.ulcerans_Agy99 GTTVRIANERAQHLDVEPVTNSDTDPDALDAEAEQVAIAEQQLLVELAEA
MLBr_01120|M.leprae_Br4923 EQLTGDEAHERVRAMLAETVDNDLWHAQRVLQAASTAAVDLSGCDALSRA
Mflv_5236|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1034|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1950|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_2989|M.marinum_M --------------------------------------------------
MAV_4077|M.avium_104 --------------------------------------------------
TH_4168|M.thermoresistible__bu --------------------------------------------------
MAB_2518|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_2033|M.ulcerans_Agy99 RDRLDAARAELADREHRAAEADRAHLAAVRAEADRREGLARLAGQVETMR
MLBr_01120|M.leprae_Br4923 LDLAAGDHAELSGTEPLLIERIEAEYRRYFTSTGRPTG------EWAVAI
Mflv_5236|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1034|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1950|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_2989|M.marinum_M --------------------------------------------------
MAV_4077|M.avium_104 --------------------------------------------------
TH_4168|M.thermoresistible__bu --------------------------------------------------
MAB_2518|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_2033|M.ulcerans_Agy99 ARVESIDDSVARLSERIEHAAARAQQTRAEFETVQARVGELDQGEVGLDE
MLBr_01120|M.leprae_Br4923 SRLSDAETAVGECAAAVAEVDDRVRRHAVLTERVAGLAQQRFAAGPRLAV
Mflv_5236|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1034|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1950|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_2989|M.marinum_M --------------------------------------------------
MAV_4077|M.avium_104 --------------------------------------------------
TH_4168|M.thermoresistible__bu --------------------------------------------------
MAB_2518|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_2033|M.ulcerans_Agy99 QHERTVAALRLAEQRLAELQVAERDAERQVASLRARIGALSVGLDRKDGA
MLBr_01120|M.leprae_Br4923 AQAAADKVAVLTRQAREAELVAAAATATNAAAVAAHTSRLRLLAEIDTRA
Mflv_5236|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1034|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1950|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_2989|M.marinum_M --------------------------------------------------
MAV_4077|M.avium_104 --------------------------------------------------
TH_4168|M.thermoresistible__bu --------------------------------------------------
MAB_2518|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_2033|M.ulcerans_Agy99 AWLARNHSDAGLFGSVAQLVKVRSGYEAAVAAVLGSAAEALAADSFGAAR
MLBr_01120|M.leprae_Br4923 VVLAATQDEAQEAVDAVSTMRADAEASDAAVEESTEALMAAQQRADIARS
Mflv_5236|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1034|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1950|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_2989|M.marinum_M --------------------------------------------------
MAV_4077|M.avium_104 --------------------------------------------------
TH_4168|M.thermoresistible__bu --------------------------------------------------
MAB_2518|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_2033|M.ulcerans_Agy99 SAVTALKQADGGRAALVLGDWPDAHDHRAAGGLPSGTLWALDLIEVPSRL
MLBr_01120|M.leprae_Br4923 TVDQLVDRQEADRLSTRLAKIDAIQGERD---------LICAELSAVTLT
Mflv_5236|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1034|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1950|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_2989|M.marinum_M --------------------------------------------------
MAV_4077|M.avium_104 --------------------------------------------------
TH_4168|M.thermoresistible__bu --------------------------------------------------
MAB_2518|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_2033|M.ulcerans_Agy99 RPAMTAMLSGVVVVNDLSEALDLVQAQPQLRAVTLDGDLVGAGWVSGGSD
MLBr_01120|M.leprae_Br4923 EQLLQRIENAAAIVDRTGEQLKLISAAVEFTATADIEISVGQQRVSLEAG
Mflv_5236|M.gilvum_PYR-GCK --------------------------------MPDPRRSEMSQDDKAAQT
Mvan_1034|M.vanbaalenii_PYR-1 ----------------------------------------MS-DDKAAQT
MSMEG_1950|M.smegmatis_MC2_155 ----------------------------------------MTQHSKSDQA
MMAR_2989|M.marinum_M --------------------------------------MADK--SGPQEA
MAV_4077|M.avium_104 --------------------------------------MGDQQ-SGPQEA
TH_4168|M.thermoresistible__bu --------------------------------------MSDNRNSGPQEA
MAB_2518|M.abscessus_ATCC_1997 --------------------------------------MTEKN-SGPQEA
MUL_2033|M.ulcerans_Agy99 RKPSTLEITSEIDKARRELAAAEVRVAELGAALSGALTDQTARQDAAEQA
MLBr_01120|M.leprae_Br4923 QSWSTTATGPTEVEVLGVLTACVIPG-------ATALDAQSKYVAAQEEL
:
Mflv_5236|M.gilvum_PYR-GCK RKSFAES-----------------VKGKAKELAGAVTGNDSLTA------
Mvan_1034|M.vanbaalenii_PYR-1 RKSFVEA-----------------LKGKAKEVAGAITGNDSLTA------
MSMEG_1950|M.smegmatis_MC2_155 RKSFIDS-----------------VKGTAKEVVGAVTGNDSLTA------
MMAR_2989|M.marinum_M VEGVVEG-----------------AKGKAKEVFGAVTGRDDVKR------
MAV_4077|M.avium_104 VKGAVEG-----------------VKGKAKEVGGTILGRDDLTE------
TH_4168|M.thermoresistible__bu IRGVVED-----------------AKGKAKEVIGTVTGREDLTR------
MAB_2518|M.abscessus_ATCC_1997 VRGVVED-----------------VKGKAKEAAGTVTGRDDLIE------
MUL_2033|M.ulcerans_Agy99 LAALNESDTAISAMYEQLGRIGQDARGAEEEWTRLLRQREELEVGRTQTL
MLBr_01120|M.leprae_Br4923 SVALADGG----------VVDLAAARCANQRRRELQSSLDQLSAALAG--
. : : :. :.:
Mflv_5236|M.gilvum_PYR-GCK ----------EGQLEQAEAKERREASRKEAEADAEAAQAQAQAREARLEG
Mvan_1034|M.vanbaalenii_PYR-1 ----------EGQLEQAEAKERRAASRNEAEADAEAAQAREKAGEAELRG
MSMEG_1950|M.smegmatis_MC2_155 ----------EGQLEKTQAKERREAGRAAAVADAEARQARAEEAQAQADG
MMAR_2989|M.marinum_M ----------EGQAQQDKADAQRDAAKK--EAEAEAARRGADVAEERQKA
MAV_4077|M.avium_104 ----------EGQAQQDKADAQRDAGKK--EAEAESARAGADAAEERQKK
TH_4168|M.thermoresistible__bu ----------EGQAQQDKADAQRDAAKK--EAEAERARANAKVDEQRQKA
MAB_2518|M.abscessus_ATCC_1997 ----------EGKAQQDKAQAQREAAQK--EAEAEKSRAEAKSDEKRQKA
MUL_2033|M.ulcerans_Agy99 EEVLELESRLRNAQESQHVQAAEPVDRQEIAAAAESARAIEVEARLTVRT
MLBr_01120|M.leprae_Br4923 ---------VCGDDQIDQLRARLEQLRDGYPGEPDLLAVDIGSARAELEA
. : . : . .: .
Mflv_5236|M.gilvum_PYR-GCK AQERAAAE---------VRAAG----VETQVSADQAAARQSVEEAGRRDV
Mvan_1034|M.vanbaalenii_PYR-1 AQQRARAE---------VQAAT----AESAVRSEQAARRQMAEESGRRDV
MSMEG_1950|M.smegmatis_MC2_155 AQKRAAVA---------AEAAV----VENDVRAQQDAQKREAEAAARRDA
MMAR_2989|M.marinum_M NQ------------------------------------------------
MAV_4077|M.avium_104 NQ------------------------------------------------
TH_4168|M.thermoresistible__bu AQRPD---------------------------------------------
MAB_2518|M.abscessus_ATCC_1997 HE------------------------------------------------
MUL_2033|M.ulcerans_Agy99 AEERANALRGRADSLRRVAAAE----REARVRAELARAVRLRSAAVAAAV
MLBr_01120|M.leprae_Br4923 AETVRAAVDFEREVCRRTAAAANCRLVETSARANFLLKKAETQRAELDQD
:
Mflv_5236|M.gilvum_PYR-GCK ARAQTQAEAKVAQESARAQAEQREEIVEAAEEYQEAAADYQEATSEAARA
Mvan_1034|M.vanbaalenii_PYR-1 AGAQSAFEADVQREAQRATAERRGRVADAAEEYHEAAADYQQKTAEAERA
MSMEG_1950|M.smegmatis_MC2_155 ERQKTQADIDAQREVQLADAAERSDVAVARQEAAEAAADHQDAVKESADV
MMAR_2989|M.marinum_M --------------------------------------------------
MAV_4077|M.avium_104 --------------------------------------------------
TH_4168|M.thermoresistible__bu --------------------------------------------------
MAB_2518|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_2033|M.ulcerans_Agy99 AESGRLLAWRLNRVVDAASQLRDELAAERQQRSAAMSAVREETSSLSARV
MLBr_01120|M.leprae_Br4923 IDQLAQQRASVSDEDLASAAEAGLRAVQIAEQRVAKLTEELVAAEPEAVT
Mflv_5236|M.gilvum_PYR-GCK EAQAERLRQRAN--DSTPT-------------------------------
Mvan_1034|M.vanbaalenii_PYR-1 EADADRLRRQAAAADDHPKLP-----------------------------
MSMEG_1950|M.smegmatis_MC2_155 RARADRMRQEADKLTEEADLPGDEK-------------------------
MMAR_2989|M.marinum_M --------------------------------------------------
MAV_4077|M.avium_104 --------------------------------------------------
TH_4168|M.thermoresistible__bu --------------------------------------------------
MAB_2518|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_2033|M.ulcerans_Agy99 ATLTDSLHRDEVANAQAALRIEQVEQMVLEQFGMAPSDLVAEYGPDVALP
MLBr_01120|M.leprae_Br4923 AELVAATAAAESLRDQHEDAAGALREISIELSVFGTEGRQGKLDTAEAER
Mflv_5236|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1034|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1950|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_2989|M.marinum_M --------------------------------------------------
MAV_4077|M.avium_104 --------------------------------------------------
TH_4168|M.thermoresistible__bu --------------------------------------------------
MAB_2518|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_2033|M.ulcerans_Agy99 PTELEMAEFEQARERGEQVVAPAPMPFDRATQQRRAKRAERELTELGRVN
MLBr_01120|M.leprae_Br4923 EHAISQHTQIGRRARAAQLLRSVMARHRDTTRLRYVEPYRTELQRLGRP-
Mflv_5236|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1034|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1950|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_2989|M.marinum_M --------------------------------------------------
MAV_4077|M.avium_104 --------------------------------------------------
TH_4168|M.thermoresistible__bu --------------------------------------------------
MAB_2518|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_2033|M.ulcerans_Agy99 PLALEEFAALEERYNFLSTQLEDVKAARKDLLDVVADVDARILQVFSDAF
MLBr_01120|M.leprae_Br4923 VFGPTFEVDIDSDLRIRSRTLDGITVPFESLSGGAKEQLGILARFAGATL
Mflv_5236|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1034|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1950|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_2989|M.marinum_M --------------------------------------------------
MAV_4077|M.avium_104 --------------------------------------------------
TH_4168|M.thermoresistible__bu --------------------------------------------------
MAB_2518|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_2033|M.ulcerans_Agy99 VDVEREFQIVFSSLFPGGEGRLRLTAPEDMLTTGIEVEARPPGKKVSRLS
MLBr_01120|M.leprae_Br4923 VAKEDNVPVVVDDALGFTDPDRLAKMGEMFDTVGAHGQVIVLTCSPDRYD
Mflv_5236|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1034|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1950|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_2989|M.marinum_M --------------------------------------------------
MAV_4077|M.avium_104 --------------------------------------------------
TH_4168|M.thermoresistible__bu --------------------------------------------------
MAB_2518|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_2033|M.ulcerans_Agy99 LLSGGEKALTAVAILVAIFRARPSPFYIMDEVEAALDDTNLRRLIGLFEM
MLBr_01120|M.leprae_Br4923 GFTGAHRIDLNV--------------------------------------
Mflv_5236|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1034|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1950|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_2989|M.marinum_M --------------------------------------------------
MAV_4077|M.avium_104 --------------------------------------------------
TH_4168|M.thermoresistible__bu --------------------------------------------------
MAB_2518|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_2033|M.ulcerans_Agy99 LRDRSQLIIITHQKPTMEVADALYGVTMQGDGITAVISQRIRGQQVDRLV
MLBr_01120|M.leprae_Br4923 --------------------------------------------------
Mflv_5236|M.gilvum_PYR-GCK ----
Mvan_1034|M.vanbaalenii_PYR-1 ----
MSMEG_1950|M.smegmatis_MC2_155 ----
MMAR_2989|M.marinum_M ----
MAV_4077|M.avium_104 ----
TH_4168|M.thermoresistible__bu ----
MAB_2518|M.abscessus_ATCC_1997 ----
MUL_2033|M.ulcerans_Agy99 ANSS
MLBr_01120|M.leprae_Br4923 ----