For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VKIVAIGAIPFSIPYTKPLRFASGEVHAAEHVLVRVHTDDGIVGVAEAPPRPFTYGETQTGIVAVIEQYF APALIGLTLTEREVAHTRMARTVGNPTAKAAIDMAMWDALGQSLRLSVSEMLGGYTDRMRVSHMLGFDDP VKMVAEAERIRETYGINTFKVKVGRRPVQLDTAVVRALRERFGDAIELYVDGNRGWSAAESLRAMREMAD LDLLFAEELCPADDVLSRRRLVGQLDMPFIADESVPTPADVTREVLGGSATAISIKTARTGFTGSTRVHH LAEGLGLDMVMGNQIDGQIGTACTVSFGTAFERTSRHAGELSNFLDMSDDLLTVPLQISDGQLHRRPGPG LGIEIDPDKLAHYRTDN
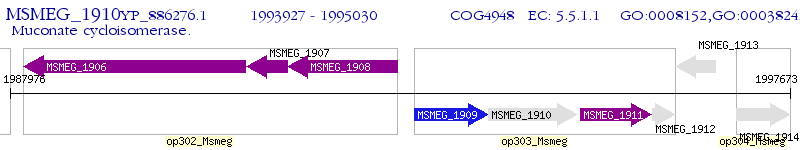
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1910 | - | - | 100% (367) | muconate cycloisomerase |
| M. smegmatis MC2 155 | MSMEG_6871 | - | 5e-27 | 28.27% (375) | isomerase |
| M. smegmatis MC2 155 | MSMEG_6177 | - | 1e-14 | 25.93% (351) | galactonate dehydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0568 | menC | 5e-07 | 27.98% (168) | O-succinylbenzoate synthase |
| M. gilvum PYR-GCK | Mflv_1357 | - | 1e-176 | 82.29% (367) | mandelate racemase/muconate lactonizing protein |
| M. tuberculosis H37Rv | Rv0553 | menC | 5e-07 | 27.98% (168) | O-succinylbenzoate synthase |
| M. leprae Br4923 | MLBr_02268 | - | 2e-05 | 29.53% (149) | O-succinylbenzoate synthase |
| M. abscessus ATCC 19977 | MAB_3935c | - | 4e-09 | 29.72% (212) | O-succinylbenzoate synthase |
| M. marinum M | MMAR_0899 | menC | 4e-06 | 26.51% (166) | muconate cycloisomerase, MenC |
| M. avium 104 | MAV_3096 | - | 5e-11 | 27.20% (239) | mandelate racemase/muconate lactonizing enzyme |
| M. thermoresistible (build 8) | TH_0152 | menC | 4e-10 | 28.44% (225) | PROBABLE MUCONATE CYCLOISOMERASE MENC (CIS,CIS-MUCONATE |
| M. ulcerans Agy99 | MUL_0652 | menC | 3e-06 | 26.51% (166) | O-succinylbenzoate synthase |
| M. vanbaalenii PYR-1 | Mvan_1017 | - | 2e-13 | 28.52% (305) | glucarate dehydratase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0899|M.marinum_M -------------------------MIPGPATLPDLLDRLH---------
MUL_0652|M.ulcerans_Agy99 -------------------------MIPGPATLPDLLDRLQ---------
Mb0568|M.bovis_AF2122/97 -------------------------MIPVLPPLEALLDRLY---------
Rv0553|M.tuberculosis_H37Rv -------------------------MIPVLPPLEALLDRLY---------
MLBr_02268|M.leprae_Br4923 -------------------------MIP---TLTELLDRLH---------
TH_0152|M.thermoresistible__bu ----------------------VGDTGPVMPTLDELLERLR---------
MAB_3935c|M.abscessus_ATCC_199 ---------------------------MELPVLDDVLDRLH---------
MSMEG_1910|M.smegmatis_MC2_155 ----MKIVAIGAIPFSIPYTKPLRFASGEVHAAEHVLVRVHTDDGIVGVA
Mflv_1357|M.gilvum_PYR-GCK ----MKITAVDAIPFAIPYTKPLKFASGEVHTAEHVLVRVHTDEGVVGVA
Mvan_1017|M.vanbaalenii_PYR-1 -MRTPVITDVTVVPIAGHDSMLLNLSGAHGPFFTRNLVIVEDSEGNTGVG
MAV_3096|M.avium_104 MTPDPAIDEVTARVYQIPTDAPEADGTLSWSSTTLVLAEVAAGGRRGIGY
* :
MMAR_0899|M.marinum_M ------------------VVALPMRVRFRGITTREVALIEGPSGWGEFGA
MUL_0652|M.ulcerans_Agy99 ------------------VVALPMRVRFRGITTREVALIEGPSGWGEFGA
Mb0568|M.bovis_AF2122/97 ------------------VVALPMRVRFRGITTREVALIEGPAGWGEFGA
Rv0553|M.tuberculosis_H37Rv ------------------VVALPMRVRFRGITTREVALIEGPAGWGEFGA
MLBr_02268|M.leprae_Br4923 ------------------VVALPMRVRFRGITTREVALIDGPAGWGEFGA
TH_0152|M.thermoresistible__bu ------------------VVALPMRVPFRGVTVREVALIEGPVAWGEFGA
MAB_3935c|M.abscessus_ATCC_199 ------------------VVALPMRVRFRGITTREVALIDGPAGWGEFGA
MSMEG_1910|M.smegmatis_MC2_155 EAPPRPFTYGETQTGIVAVIEQYFAPALIGLTLTEREVAHTRMAR-TVGN
Mflv_1357|M.gilvum_PYR-GCK EAPPRPFTYGETQDGILAVIRQIFAPQIVGLTLTEREVVNARLAR-TVGN
Mvan_1017|M.vanbaalenii_PYR-1 EVPGGEPIRRTLQDARGIVTGRSIGDYHAVLNEMRRTFADRDAGGRGAQT
MAV_3096|M.avium_104 TYADGACAALITG----ALAGSVTGHSALDVPARWQAMVRAMRNNGRPGL
: : .
MMAR_0899|M.marinum_M FLEYQ------PPEAAAWLASAIDAAYGQPPPVR--RGRIPINAT-----
MUL_0652|M.ulcerans_Agy99 FLEYQ------PPEAAAWLASAIDAAYGQPPPVR--RGRIPINAT-----
Mb0568|M.bovis_AF2122/97 FVEYQ------SAQACAWLASAIETAYCAPPPVR--RDRVPINAT-----
Rv0553|M.tuberculosis_H37Rv FVEYQ------SAQACAWLASAIETAYCAPPPVR--RDRVPINAT-----
MLBr_02268|M.leprae_Br4923 FLEYQ------PLEASAWLVAGIEAAYGEPPEGR--RDRIPINAT-----
TH_0152|M.thermoresistible__bu FPEYG------PAEAAAWLAAGIEAAFKSPPLIR--RHRIPINAT-----
MAB_3935c|M.abscessus_ATCC_199 FPEYQ------PAEASAWLSAAIEAAYRALPTPR--RDRIPVNAT-----
MSMEG_1910|M.smegmatis_MC2_155 PTAKA------AIDMAMWDALGQSLRLSVSEMLGGYTDRMRVSHM-----
Mflv_1357|M.gilvum_PYR-GCK PTAKA------AVDMAIWDALGRSLDLSVSALLGGYGDHMRVSHM-----
Mvan_1017|M.vanbaalenii_PYR-1 FDLRVTVHAVTAIESALMDLLGQHLEVPVAALLGDGQQRSRVQALGYLFF
MAV_3096|M.avium_104 VSCAIS-----AVDTALWDLKGKLLGLPVCRLLGMAHDSVPIYGSG----
. : . . :
MMAR_0899|M.marinum_M -----------------------------VPAVPPAQVPELLARFP---G
MUL_0652|M.ulcerans_Agy99 -----------------------------VPAVPPAQVPELLARFP---G
Mb0568|M.bovis_AF2122/97 -----------------------------VPAVAAAQVGEVLARFP---G
Rv0553|M.tuberculosis_H37Rv -----------------------------VPAVAAAQVGEVLARFP---G
MLBr_02268|M.leprae_Br4923 -----------------------------VPAVSATQVPEVLARFP---G
TH_0152|M.thermoresistible__bu -----------------------------VPAVPAQRVPEVLSRFP---G
MAB_3935c|M.abscessus_ATCC_199 -----------------------------VPAVTAADVPEVLSRFP---G
MSMEG_1910|M.smegmatis_MC2_155 -----------------------------LGFDDPVKMVAEAERIRETYG
Mflv_1357|M.gilvum_PYR-GCK -----------------------------LGFDTPDTMVAEAERIRDEYG
Mvan_1017|M.vanbaalenii_PYR-1 VGDRTRTDLAYRSPSDEEPGADDWFTVRHEEAMTPDAVVRLAEAARARYG
MAV_3096|M.avium_104 ----------------------------GFTSYDDNATRAQLERWVHEWR
MMAR_0899|M.marinum_M ARTAKVKVAE---PGQTLASDVERVNAVR-ALVPTVRVDANGGWSVDQAA
MUL_0652|M.ulcerans_Agy99 ARTAKVKVAE---PGQTLASDVERVNAVR-ALVPTVRVDANGGWSVDQAA
Mb0568|M.bovis_AF2122/97 ARTAKVKVAE---PGQSLADDIERVNAVR-ELVPMVRVDANGGWGVAEAV
Rv0553|M.tuberculosis_H37Rv ARTAKVKVAE---PGQSLADDIERVNAVR-ELVPMVRVDANGGWGVAEAV
MLBr_02268|M.leprae_Br4923 VRTAKVKVAE---PGQNLADDVDRVNAVR-ELVETVRVDANGGWSVEAAA
TH_0152|M.thermoresistible__bu ARTAKVKVAE---PGQTLTDDVARVNAVR-ELVPTVRVDVNGAWTVDEAV
MAB_3935c|M.abscessus_ATCC_199 AQTAKVKVAE---PGQTLDEDVARVEAVR-ARIRTVRVDANGGWTVAQAV
MSMEG_1910|M.smegmatis_MC2_155 INTFKVKVGR---RPVQLDTAVVRALRERFGDAIELYVDGNRGWSAAESL
Mflv_1357|M.gilvum_PYR-GCK ISTFKVKVGR---RPVTLDTAVVRALRERFGDTVELYVDGNRGWTASESL
Mvan_1017|M.vanbaalenii_PYR-1 FADFKLKGGV---LPAADEAKAVIALAER-FPDSRITLDPNGGWLLADAI
MAV_3096|M.avium_104 IPRVKIKIGESWGSHERRDLDRIALARTVIGPDTELYVDANGGYRRKQAI
*:* . : :* * .: :
MMAR_0899|M.marinum_M QAAVALTADGPLEYLEQPCATVGELAELRRRIE------VPIAADEAIRK
MUL_0652|M.ulcerans_Agy99 QAAVALTADGPLEYLEQPCATVGELAELRRRIE------VPIAADEAIRK
Mb0568|M.bovis_AF2122/97 AAAAALTADGPLEYLEQPCATVAELAELRRRVD------VPIAADESIRK
Rv0553|M.tuberculosis_H37Rv AAAAALTADGPLEYLEQPCATVAELAELRRRVD------VPIAADESIRK
MLBr_02268|M.leprae_Br4923 QAAVALTADGPLEYLEQPCATVAELAALRRRVD------IPIAADESIRK
TH_0152|M.thermoresistible__bu KAAAALTADGPLEYLEQPCATIAELAELRRRID------VPIAADESIRK
MAB_3935c|M.abscessus_ATCC_199 HALTALTRDGALEYAEQPCATVEELAELRRRLPG-----VPIAADESIRR
MSMEG_1910|M.smegmatis_MC2_155 RAMREM-ADLDLLFAEELCPADDVLSRRRLVGQLD----MPFIADESVPT
Mflv_1357|M.gilvum_PYR-GCK RALKDM-ADLDLLFAEELCPADDVLGRRWLVEHTE----IPFIADESVPT
Mvan_1017|M.vanbaalenii_PYR-1 RTCREL--KDVLAYAEDPVGPEDGFSGREVMAEFKRATGLPTATNMIATD
MAV_3096|M.avium_104 RMAHAM-AGHDVTWFEEPVSSDDLAGLREVRDQVS----ADVTAGEYGYD
: : : *: . . :.
MMAR_0899|M.marinum_M AEDPLAVVRAGAADVAVLKVAPLG--GVWALLDIAAQIDIPVVISSALDS
MUL_0652|M.ulcerans_Agy99 AEDPLAVVRAGAADVAVLKVAPLG--GVWALLDIAAQIDIPVVISSALDS
Mb0568|M.bovis_AF2122/97 AEDPLAVVRAQAADIAVLKVAPLG--GISALLDIAARIAVPVVVSSALDS
Rv0553|M.tuberculosis_H37Rv AEDPLAVVRAQAADIAVLKVAPLG--GISALLDIAARIAVPVVVSSALDS
MLBr_02268|M.leprae_Br4923 ADDPLAVVRSHAADVAVLKVAPLG--GIASFLAIAAQIDIPVVVSSALDS
TH_0152|M.thermoresistible__bu ADDPLRVVRAGAADIAVLKVAPLG--GAYRLLEIATAIDIPIVVSSALDT
MAB_3935c|M.abscessus_ATCC_199 ASDPLRVAQARAADIAVLKVAPLG--GVHALLEIAARIGMPAVISSALDS
MSMEG_1910|M.smegmatis_MC2_155 PADVTREVLGGSATAISIKTARTGFTGSTRVHHLAEGLGLDMVMGNQIDG
Mflv_1357|M.gilvum_PYR-GCK PADVTREVLGGSATAISIKTARTGFTGSRRVHHLAEGLGLEVVMGNQIDG
Mvan_1017|M.vanbaalenii_PYR-1 WREMGHAIRSGAVDIPLADPHFWTMTGSVRVAQLCDAWGLTWGSHSNNHF
MAV_3096|M.avium_104 LAYFHRMLAADAVDCLQIDVTRCG-----GITDWLRAAAVAAAHNVDVSG
. :. . . :
MMAR_0899|M.marinum_M AVGIAAGLSAAAALPQLQHACG-------LGTGGLFVEDVAEPAIPVDGA
MUL_0652|M.ulcerans_Agy99 AVGIAAGLSAAAALPQLQHACG-------LGTGGLFVEDVAEPAIPVDGA
Mb0568|M.bovis_AF2122/97 AVGIAAGLTAAAALPELDHACG-------LGTGGLFEEDVAEPAAPVDGF
Rv0553|M.tuberculosis_H37Rv AVGIAAGLTAAAALPELDHACG-------LGTGGLFEEDVAEPAAPVDGF
MLBr_02268|M.leprae_Br4923 VVGITAGLIAAAALPELDYACG-------LGTGRLFVADVAEPMLPVDGF
TH_0152|M.thermoresistible__bu AVGIGRGLLAAAALPQLNHACG-------LGTGRLFVEDVAEPLELVDGH
MAB_3935c|M.abscessus_ATCC_199 AVGIGIGLRAAAALPELDHACG-------LGTGGFFLADVADVPPVVDGH
MSMEG_1910|M.smegmatis_MC2_155 QIGTACTVSFGTAFERTSRHAG-------ELSN---FLDMSDDLLTVPLQ
Mflv_1357|M.gilvum_PYR-GCK QLGTACAVAFGAAFELTSRRAG-------ELSN---FLDMSDDLLTHPLR
Mvan_1017|M.vanbaalenii_PYR-1 DVSLAMFTHVAAAAPGDITAIDTHWIWQDGQAITTNPYPIVDGYLSVPDA
MAV_3096|M.avium_104 HCAPNLHAHVATAVANLRHLEY------------FHDHHRIERMFFDGAL
. .:* :
MMAR_0899|M.marinum_M LAPQRVVPDPARLRALGAAPDRRQWWIDRIKACYPLLYRRSGD-------
MUL_0652|M.ulcerans_Agy99 LAPQRVAPDPARLRALGAAPDRRQWWIDRIKACYPLLYRRSGD-------
Mb0568|M.bovis_AF2122/97 LAVARTTPDPARLQALGAPPQRRQWWIDRVKACYSLLVPSFG--------
Rv0553|M.tuberculosis_H37Rv LAVARTTPDPARLQALGAPPQRRQWWIDRVKACYSLLVPSFG--------
MLBr_02268|M.leprae_Br4923 LPVGPVTPDPARLQALGTPPARRQWWIDRVKTCYSLLVPCAGDQSGLRRP
TH_0152|M.thermoresistible__bu LPAAPVRPDPQRVLELAAPPDRRRWWMDRTRECFALLET-----------
MAB_3935c|M.abscessus_ATCC_199 LPVTSCTPDPARLAALAVPADRLDRWRRRVADCHALLNR-----------
MSMEG_1910|M.smegmatis_MC2_155 ISDGQLHRRPGPGLGIEIDPDKLAHYRTDN--------------------
Mflv_1357|M.gilvum_PYR-GCK IRDGRLHVPPGPGLGIEIDPDKLDHYRTDH--------------------
Mvan_1017|M.vanbaalenii_PYR-1 PGLGVTLDEPAVQAAHALYQREGLGGRDDAVAMQYLIPGWTFDGKRPALD
MAV_3096|M.avium_104 SPQGGVLRPDQQRPGFGLEFKQRDADRYRMR-------------------
.
MMAR_0899|M.marinum_M ---
MUL_0652|M.ulcerans_Agy99 ---
Mb0568|M.bovis_AF2122/97 ---
Rv0553|M.tuberculosis_H37Rv ---
MLBr_02268|M.leprae_Br4923 RDR
TH_0152|M.thermoresistible__bu ---
MAB_3935c|M.abscessus_ATCC_199 ---
MSMEG_1910|M.smegmatis_MC2_155 ---
Mflv_1357|M.gilvum_PYR-GCK ---
Mvan_1017|M.vanbaalenii_PYR-1 RG-
MAV_3096|M.avium_104 ---