For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKIKSVQTYTVPPRWLFLRIETDEGLVGWGEPVLEGRAATVAAAVEELSDYLIGQDPGQIEDLWTVMYRG GFYRGGGIHMSALAGIDQALWDLKGKTLGVPTHELLGGRVRDRIKVYSWIGGDRPSETAKAAREVVDRGF TAVKMNGTEELQYVDTWDKVDRAVASVAAVRDAVGPNIGIGVDFHGRVHKPMAKVLLRELEPYRLMFVEE PVLSEHVDGFVDVLRQSPIPIALGERLFSRWDAKTILASGAVDIIQPDPSHCGGITEARKIAHMAEAYDV ALALHCPLGPIALAACLQIDAGCYNATIQEQSLGIHYNTTNDLLDYLADPGVFTYADGQVQIPTGPGLGI EINEEYVAERAAEGHRWRNPVWRHADGSFAEW
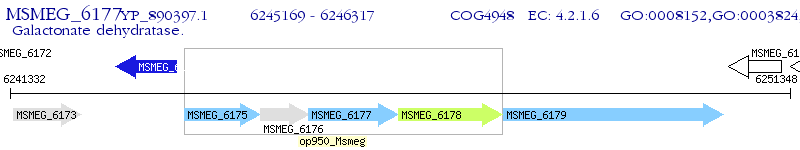
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6177 | - | - | 100% (382) | galactonate dehydratase |
| M. smegmatis MC2 155 | MSMEG_2909 | - | 3e-51 | 31.63% (411) | starvation-sensing protein RspA |
| M. smegmatis MC2 155 | MSMEG_6871 | - | 3e-32 | 29.86% (355) | isomerase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1357 | - | 7e-15 | 23.69% (363) | mandelate racemase/muconate lactonizing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0864 | - | 3e-07 | 27.78% (216) | mandelate racemase |
| M. avium 104 | MAV_3096 | - | 2e-20 | 27.65% (264) | mandelate racemase/muconate lactonizing enzyme |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1029 | - | 2e-20 | 25.48% (365) | mandelate racemase/muconate lactonizing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1357|M.gilvum_PYR-GCK ---MKITAVDAIPFAIPYTKPLKFASGEVHTAEHVLVRVHTDEGVVGVAE
Mvan_1029|M.vanbaalenii_PYR-1 MAPIRVTGARITPVAFADPPLLNTVGVHQPYALRAIIQLDTDAGLVGLGE
MSMEG_6177|M.smegmatis_MC2_155 -------------------MKIKSVQTYTVPPRWLFLRIETDEGLVGWGE
MAV_3096|M.avium_104 MTPDPAIDEVTARVYQIPTDAPEADGTLSWSSTTLVLAEVAAGGRRGIGY
MMAR_0864|M.marinum_M MRIVAILERPVRLLGGPANAVVNFSQHTVSLVAVVSDVIRHGKPLTGVAF
: * .
Mflv_1357|M.gilvum_PYR-GCK APPRPFTYGETQDGIL-----AVIR-QIFAP--QIVGLTLTEREVVNARL
Mvan_1029|M.vanbaalenii_PYR-1 TYADTRHLARLRAAAE-----AITGLDVFAL--NRIRASIGSRLEGDTTA
MSMEG_6177|M.smegmatis_MC2_155 PVLEG-RAATVAAAVE-----ELSDYLIGQD--PGQIEDLWTVMYRGGFY
MAV_3096|M.avium_104 TYADGACAALITGALAG----SVTGHSALDV--PARWQAMVRAMRNNGRP
MMAR_0864|M.marinum_M NSIGRFAQSGILRDRMIPRVLAAPEDALLDQRGHLDPAAVLRHAVCDEKP
. : .
Mflv_1357|M.gilvum_PYR-GCK ARTVGNPTAKAAVD-------MAIWDALGRSLD----LSVSALLGGYG-D
Mvan_1029|M.vanbaalenii_PYR-1 VGTAGMITSASVVDQVLSPFEVACLDVQGQSLG----RPVSDLLGGAVRD
MSMEG_6177|M.smegmatis_MC2_155 RGGGIHMSALAGID-------QALWDLKGKTLG----VPTHELLGGRVRD
MAV_3096|M.avium_104 ---GLVSCAISAVD-------TALWDLKGKLLG----LPVCRLL-GMAHD
MMAR_0864|M.marinum_M GGHGDRAGAAAAVE-------LACWDLAAKLNDEPAYATIARHFGRDPAP
: : :: * * .: . . :
Mflv_1357|M.gilvum_PYR-GCK HMRVSHMLGFD----------------TPDTMVAEAERIRDEYGISTFKV
Mvan_1029|M.vanbaalenii_PYR-1 AVPFSAYLFYKWAGHPNAEPDRFGEAMDPNGLVAQARRIIDEYGFTAIKL
MSMEG_6177|M.smegmatis_MC2_155 RIKVYSWIGGDRPSE---------TAKAAREVVDRGFTAVKMNGTEELQY
MAV_3096|M.avium_104 SVPIYGSGGFTS--------------YDDNATRAQLERWVHEWRIPRVKI
MMAR_0864|M.marinum_M LVSVYAAGGYYYPGG------------GLNALRDEMRRYLDLG-YDAVKM
: . . . .:
Mflv_1357|M.gilvum_PYR-GCK KVGR---RPVTLDTAVVRALRERFGDTVELYVDGNRGWTASESLRALKDM
Mvan_1029|M.vanbaalenii_PYR-1 KGGV---FPPEEEMAAIEALSRNFPG-LPLRLDPNAAWTPHTAVKVASGL
MSMEG_6177|M.smegmatis_MC2_155 VDTW---DKVDRAVASVAAVRDAVGPNIGIGVDFHGRVHKPMAKVLLREL
MAV_3096|M.avium_104 KIGESWGSHERRDLDRIALARTVIGPDTELYVDANGGYRRKQAIRMAHAM
MMAR_0864|M.marinum_M KIGG---VAQRDDLARIEAVIDVVGA-DRVAVDANGRFDRAAATRWAATL
: . : :* : : :
Mflv_1357|M.gilvum_PYR-GCK ADLDLLFAEELCPADDVLGRRWLVEHTEIPFIADESVPTPADVTREVLG-
Mvan_1029|M.vanbaalenii_PYR-1 AGILEYLED---PTPGLAGMAEVAQQAPMPLATNMCVVAFDQLAPAVTKN
MSMEG_6177|M.smegmatis_MC2_155 EPYRLMFVEEPVLSEHVDGFVDVLRQSPIPIALGERLFSRWDAKTILASG
MAV_3096|M.avium_104 AGHDVTWFEEPVSSDDLAGLREVRDQVSADVTAGEYGYDLAYFHRMLAAD
MMAR_0864|M.marinum_M APYGLRWYEEPGDPLDFELNRAVTGCYAGTVATGENLFSVPDVTNLVRYG
: . . : . . :
Mflv_1357|M.gilvum_PYR-GCK ----GSATAISIKTARTGFTGSRRVHHLAEGLGLEVVMGNQIDGQLGTAC
Mvan_1029|M.vanbaalenii_PYR-1 ----AVRVVLSDHHYWGGLQRSRLLAGICDTFGLGLSMHSNSHLGISLAA
MSMEG_6177|M.smegmatis_MC2_155 ----AVDIIQPDPSHCGGITEARKIAHMAEAYDVALALHCPLG-PIALAA
MAV_3096|M.avium_104 ----AVDCLQIDVTRCGGITDWLRAAAVAAAHNVDVSGHCAPNLHAHVAT
MMAR_0864|M.marinum_M GMRAGRDIFQMDAGLSYGLGEYVRMLELIEAHGFDRRCAYPHGGHMINLH
. *: : ..
Mflv_1357|M.gilvum_PYR-GCK AVAFGAAFELTSRRAGELSNFLDMSDDLLT-----HPLRIRDGRLHVPPG
Mvan_1029|M.vanbaalenii_PYR-1 MVHLAGATPNLT-YACDTHWPWKTEDVVKD-----GALAFVDGAVPVPTS
MSMEG_6177|M.smegmatis_MC2_155 CLQIDAGCYNATIQEQSLGIHYNTTNDLLDYLADPGVFTYADGQVQIPTG
MAV_3096|M.avium_104 AVAN-------LRHLEYFHDHHRIERMFFD-----GALSPQGGVLRPDQQ
MMAR_0864|M.marinum_M IAAGLG-----LGGCESYPGVFQPFGGYCD------GCVLRDGQIVPTDA
.* :
Mflv_1357|M.gilvum_PYR-GCK -PGLGIEIDPDKLDHYRTDH----------------------------
Mvan_1029|M.vanbaalenii_PYR-1 -PGLGVEIDDDALDALHEQYVRCGIRDRDDTGYMRTVDPSFEPAGPRW
MSMEG_6177|M.smegmatis_MC2_155 -PGLGIEINEEYVAERAAEG------HRWRNPVWRHADGSFAEW----
MAV_3096|M.avium_104 RPGFGLEFKQRDADRYRMR-----------------------------
MMAR_0864|M.marinum_M -PGFGLERKPGLGELISELTR---------------------------
**:*:* .