For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DAYNDLRDIQIPTRKAAEVTGMHRSTATRRAKPAPAPADRAPRPVPVNKLTESECARVVEVLNSARFVDA APMQVWATLLDEGSYLCSVSTMYRVLNANKLVKERRRLARHRKAVCPELVATAPRQVYSWDITKLAGPVK GVYYDAYVMIDIYSRYIVGVHVHARECGQLAKEMMEQIFGTYGIPHVVHADRGTSMTSNTVAGLLSDLGV IRSHSRPKVSNDNPYSESWFKTLKYAPAFPERFESIHHARDFMDRFVQWYNHEHRHSGIGLHTPADVFYG LAGGKDTQRRAVLADARARHPRRFGRDTAPKVIDLPESAAINPPKPAEQETETEAA*
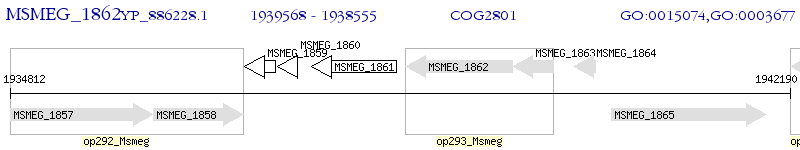
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1862 | - | - | 100% (337) | transposase |
| M. smegmatis MC2 155 | MSMEG_4133 | - | 0.0 | 99.41% (337) | transposase |
| M. smegmatis MC2 155 | MSMEG_1726 | - | 0.0 | 99.41% (337) | transposase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3460c | - | 8e-73 | 52.80% (286) | transposase |
| M. gilvum PYR-GCK | Mflv_4078 | - | 5e-49 | 37.67% (300) | integrase catalytic subunit |
| M. tuberculosis H37Rv | Rv3430c | - | 2e-73 | 53.15% (286) | transposase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2088 | - | 4e-07 | 24.23% (194) | transposase |
| M. marinum M | MMAR_4846 | - | 2e-07 | 28.18% (181) | transposase, ISMyma01_aa2 |
| M. avium 104 | MAV_1454 | - | 5e-09 | 29.14% (175) | transposase |
| M. thermoresistible (build 8) | TH_4372 | - | 3e-10 | 33.33% (90) | PUTATIVE Integrase, catalytic region |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0306 | - | 5e-47 | 37.37% (289) | integrase catalytic subunit |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3460c|M.bovis_AF2122/97 MIDTAIEEMIPLIGVRAACAATGRAPASYYRAHSKRLSAQSDTFTSTAVT
Rv3430c|M.tuberculosis_H37Rv MIDTAIEEMIPLIGVRAACAATGRAPASYYRAHSKRLSAQSDTFTSTAVT
MSMEG_1862|M.smegmatis_MC2_155 -MDAYNDLRDIQIPTRKAAEVTG-----MHRSTATRRAKP-----APAPA
Mflv_4078|M.gilvum_PYR-GCK ------------MTAAIGSQRQALAMIDLSRSTWHYRTNP-----RPPVS
Mvan_0306|M.vanbaalenii_PYR-1 -------------------------MIELSRSTWHYRSHP-----RPRVA
MAV_1454|M.avium_104 ----------------------------MALSTYYDTKAR--------VP
MAB_2088|M.abscessus_ATCC_1997 ---------------------MTRRVLKLARQPYYRWRAN--------PI
TH_4372|M.thermoresistible__bu ----VSRFELIAAECAHHDVSKLAELLGVSRSGYYAWAAR-----QRRVE
MMAR_4846|M.marinum_M -------------------MLGVSERFACRVTGQHRATQR-------HEP
Mb3460c|M.bovis_AF2122/97 DPSGPRESAQPRALSAAEREHVLAVLNSQRFADMAPAVVYATLLDEGIYL
Rv3430c|M.tuberculosis_H37Rv DPSGPRESAQPRALSAAEREHVLAVLNSQRFADMAPAVVYATLLDEGIYL
MSMEG_1862|M.smegmatis_MC2_155 DR-APR-PVPVNKLTESECARVVEVLNSARFVDAAPMQVWATLLDEGSYL
Mflv_4078|M.gilvum_PYR-GCK DPLPQKHRAYPSRIDEADRAVIRDKILAGWEAGSSVDHAFAASWDEGVML
Mvan_0306|M.vanbaalenii_PYR-1 NPMPHKDRAYPSRIGVADRAVIAEQIIAGWLAGVSVDQSFATMWDAGVLL
MAV_1454|M.avium_104 SARALRDAVLGPALCQLWKDNYCVYG-ARKLWKTARREGHDVGRDQVARL
MAB_2088|M.abscessus_ATCC_1997 TDAELVEAYRANALFDAHGE--DPEFGYRYLVEEAREAGEPMAERTAWRI
TH_4372|M.thermoresistible__bu LSPRQQWRRDLEVKILAHWQASGRTYGSPRITADLHAEGVGVSENTVAKI
MMAR_4846|M.marinum_M VAGTPADPDAALREWLRDYANKHPRRGFRPAYHDARGEGWEVNHKKVQRL
:
Mb3460c|M.bovis_AF2122/97 CSESTMYRLLRERGQTGDR----RRQATHPAAVKPELVAHQPNSVWSWDI
Rv3430c|M.tuberculosis_H37Rv CSESTMYRLLRERGQTGDR----RRQATHPAAVKPELVAHQPNSVWSWDI
MSMEG_1862|M.smegmatis_MC2_155 CSVSTMYRVLNANKLVKER----RRLARHRKAVCPELVATAPRQVYSWDI
Mflv_4078|M.gilvum_PYR-GCK ASRRSWWRIAAAIEDQSARPVAPTRSTSKTPRQAPVLKATGPQQIWSWDI
Mvan_0306|M.vanbaalenii_PYR-1 ASRRSWWRIAAVI-DQSLRPVAPTRATNKTPRSAPVLKATGPQQIWSWDI
MAV_1454|M.avium_104 MRAAGIEGVRRGKRVRTTKP--DPAAARHPDLVKRNFTATAPNQLWVTDL
MAB_2088|M.abscessus_ATCC_1997 CSQQQLWSVFGKKRGKNGK----PGPPVHDDLVQRDFTAETPNMLWLSDI
TH_4372|M.thermoresistible__bu MAEMGIEGISPRTFKVKTTQ-VDPTASFPPDRVGRAFDQGRLDAVWTSDI
MMAR_4846|M.marinum_M WREEGLRVPQRRKRKRHGTS-----------TAPPEVVADAPDRVWAVDF
. :: *:
Mb3460c|M.bovis_AF2122/97 TKLRGPAKWSYYYLYVILDIFSRYVVGWMVASRESKVLAERLIAQTLAAQ
Rv3430c|M.tuberculosis_H37Rv TKLRGPAKWSYYYLYVILDIFSRYVVGWMVASRESKVLAERLIAQTLAAQ
MSMEG_1862|M.smegmatis_MC2_155 TKLAGPVKGVYYDAYVMIDIYSRYIVGVHVHARECGQLAKEMMEQIFGTY
Mflv_4078|M.gilvum_PYR-GCK TDLRSPWRGVAFKAYSILDIYSRKIVGWRVEERECDDLAVDMFEAAFAGH
Mvan_0306|M.vanbaalenii_PYR-1 TDLHSPWRGVSFKTYSIVDIFSRKIVGWRVEERECDDLAVEMFKTAIAEH
MAV_1454|M.avium_104 TFVP-TWAGVAY-VCFIIDAYSRMIVGWRVASHMRTSMVLDALEMARWSR
MAB_2088|M.abscessus_ATCC_1997 TEHY--TGDGKLYLCAIKDAFSNRIVGYSIDSRMKSRLATAALRNAVGRR
TH_4372|M.thermoresistible__bu TYLT--CGEGDAYLCAIRDEHSRRVLGWSLADHMRTELVEAAVDAAVFTR
MMAR_4846|M.marinum_M QFDVT-TDGRPIKIVSIIDEHTRECLGGMVERSITGDHLTEELGRIAAGR
: * .:. :* : .
Mb3460c|M.bovis_AF2122/97 HISADQLTLHADRGSSMSSKPVALLLADLGVTKSHSHPHTSNDNPLSEAQ
Rv3430c|M.tuberculosis_H37Rv HISADQLTLHADRGSSMSSKPVALLLADLGVTKSHSRPHTSNDNPLSEAQ
MSMEG_1862|M.smegmatis_MC2_155 GIPH---VVHADRGTSMTSNTVAGLLSDLGVIRSHSRPKVSNDNPYSESW
Mflv_4078|M.gilvum_PYR-GCK GLPA---VVHADSGPAMRSTVLKDLLAGLKIGQTHNRPRVSNDNPFSESE
Mvan_0306|M.vanbaalenii_PYR-1 GPPA---VVHADSGPAMRSTVLKDLLAGLGIGQTHIRPHVSNDNPFSESE
MAV_1454|M.avium_104 GNMLPGLRCHSDAGSQFTSIRYGERLAEIGAVPSIGTVGDSFDNALAETV
MAB_2088|M.abscessus_ATCC_1997 GVVAG-CVVHTDRGSQFRSRKFVQALHHHGMVGSMGRVGAAGDNAAMESF
TH_4372|M.thermoresistible__bu AGHVAGTVMHSDRGSQFTSYDMAQACTRHGLLRSMGATGICWDNAGSESL
MMAR_4846|M.marinum_M GALP--AVLRCDNGPELACSAMADWAEGQ-VGLHFIPPGEPWRNGYVESF
: * *. : . * *:
Mb3460c|M.bovis_AF2122/97 ----FKTLKYRPDFPKRFESIEAARVHCDRFFG-WYNHEHKHSGIGLHTP
Rv3430c|M.tuberculosis_H37Rv ----FKTLKYRPDFPKRFESIEAARVHCDRFFG-WYNHEHKHSGIGLHTP
MSMEG_1862|M.smegmatis_MC2_155 ----FKTLKYAPAFPERFESIHHARDFMDRFVQ-WYNHEHRHSGIGLHTP
Mflv_4078|M.gilvum_PYR-GCK ----FRTMKYRPNYPGVFTDLDSARAWVSAYVS-WYNQHHRHSGVELFTP
Mvan_0306|M.vanbaalenii_PYR-1 ----FRTMKYRPNYPGVFDDLNTAHAWVQAYVR-WYNQHHRHSGIALFTP
MAV_1454|M.avium_104 NGYYKAELIYGPARTGPWKTVEDVELATLSWVY-WHNTSRLHSYLGDIPP
MAB_2088|M.abscessus_ATCC_1997 ----FSLLQKNVLDRRRWATREELRIAIVTWIERTYHRRRRQGALGRLTP
TH_4372|M.thermoresistible__bu ----WSTVKHEYYKRHAFTTYANLTAGLDNYIR-FYNHERRHSSLGMISP
MMAR_4846|M.marinum_M N----SRIRDECLNINSFWSLAQARVVIGDWKH-EYNHHRRHSSLGYLPP
: : : :: : :. : .*
Mb3460c|M.bovis_AF2122/97 ADVHYGRADQIRRHRATVLDTAYRDHLERIRSQTTRATRATGLQRDQPTT
Rv3430c|M.tuberculosis_H37Rv ADVHYGRADQIRRHRATVLDTAYRDHLERIRSQTTRATRATGLQRDQPTT
MSMEG_1862|M.smegmatis_MC2_155 ADVFYGLAGGKDTQRRAVLADARARHPRRFGRDT--APKVIDLP------
Mflv_4078|M.gilvum_PYR-GCK DSVHDGTWAQLWDRRDTALQAYYDAHPERFRNRPRTKSPS----------
Mvan_0306|M.vanbaalenii_PYR-1 DEVHNGSWTRQWMHRDHILQAYYHAHPERFRARPHTKSPA----------
MAV_1454|M.avium_104 AEFEAAFYDAYRTDQPLIGIQ-----------------------------
MAB_2088|M.abscessus_ATCC_1997 VEYEAIMTTTASQAA-----------------------------------
TH_4372|M.thermoresistible__bu IDFDLSLDILLSTQV-----------------------------------
MMAR_4846|M.marinum_M ARYAAACTHR----------------------------------------
Mb3460c|M.bovis_AF2122/97 EGGPADSINPRKSCLRNVDRFRPGLLDLPAPAPVDLRRLLPSGQIR
Rv3430c|M.tuberculosis_H37Rv EGGPADSINPRKSCLRNVDRFRPGLLDLPAPAPVDLRRLLPSGQIR
MSMEG_1862|M.smegmatis_MC2_155 ---ESAAINPPK----------------PAEQETETEAA-------
Mflv_4078|M.gilvum_PYR-GCK ---PVVGINLPT-----------------ENDPDRLQAA-------
Mvan_0306|M.vanbaalenii_PYR-1 ---AIVGINLPT-----------------EKDPNRLHAA-------
MAV_1454|M.avium_104 ----------------------------------------------
MAB_2088|M.abscessus_ATCC_1997 ----------------------------------------------
TH_4372|M.thermoresistible__bu ----------------------------------------------
MMAR_4846|M.marinum_M ----------------------------------------------