For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ADFVIVHGSWHDGTLLEPVAAAIRGLGHRAYAPTVAGHGHGADTDVSIDDGVQSVIDYCRTRDLREIVLV GHSLGGTIIARVAEEIPDRITRLIFWSAFVPRPGRSITEEVEQPSTPAAEKQAPATGSSETLSLQVWRDV FVPDVDPEQAATWHALLSPEPRRPKTERLDLRRFYRSTLPMHYIDAVDDRALPRGLDREAMIERLKNVRV HRVRGGHEVLFTDPAGIAAVIVEAGVS*
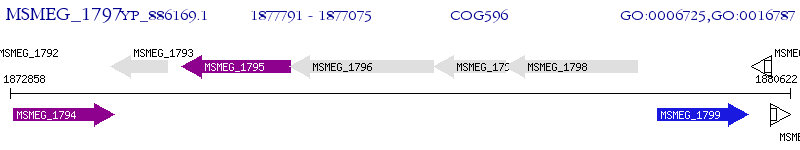
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1797 | - | - | 100% (238) | salicylate esterase |
| M. smegmatis MC2 155 | MSMEG_3842 | - | 1e-13 | 35.71% (112) | putative esterase |
| M. smegmatis MC2 155 | MSMEG_6710 | - | 7e-13 | 28.70% (230) | hydrolase, alpha/beta fold family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0079c | - | 1e-06 | 33.00% (100) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_2476 | - | 2e-14 | 25.60% (250) | esterase EstC, putative |
| M. tuberculosis H37Rv | Rv0077c | - | 1e-06 | 33.00% (100) | oxidoreductase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0768 | - | 1e-09 | 26.58% (237) | hypothetical protein MAB_0768 |
| M. marinum M | MMAR_0223 | - | 5e-08 | 29.19% (185) | oxidoreductase |
| M. avium 104 | MAV_2604 | - | 6e-15 | 30.25% (238) | hydrolase, alpha/beta fold family protein, putative |
| M. thermoresistible (build 8) | TH_1085 | - | 1e-11 | 34.55% (110) | putative esterase |
| M. ulcerans Agy99 | MUL_4891 | - | 6e-08 | 29.19% (185) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_3363 | - | 2e-13 | 34.21% (114) | hypothetical protein Mvan_3363 |
CLUSTAL 2.0.9 multiple sequence alignment
TH_1085|M.thermoresistible__bu --------------------MSTFVLVPGACHGAWWYDDLADRLRAH--G
Mvan_3363|M.vanbaalenii_PYR-1 --------------------MSSYVLIPGMCHGAWCFDEVAASLRSA--G
Mb0079c|M.bovis_AF2122/97 MSTIDISAGTIHYEATGPETGRPVVFVHGYMMGGQLWRRVSERLAGR--G
Rv0077c|M.tuberculosis_H37Rv MSTIDISAGTIHYEATGPETGRPVVFVHGYMMGGQLWRRVSERLAGR--G
MMAR_0223|M.marinum_M MPTIDINAGTIHYEVTGPEHGRPVMFVHGYLMGGQLWRQVSERLAKR--G
MUL_4891|M.ulcerans_Agy99 MPTIDINAGTIHYEITGPEHGRPVMFVHGYLMGGQLWRQVSERLAEW--G
MSMEG_1797|M.smegmatis_MC2_155 --------------------MADFVIVHGSWHDGTLLEPVAAAIRGL--G
MAV_2604|M.avium_104 ------------------MALPDLVLVHGGEHAGDCWDLVIAELRRQEPG
Mflv_2476|M.gilvum_PYR-GCK ---------------------MRFILVHGGFHAAWCWERTAAALRAL--G
MAB_0768|M.abscessus_ATCC_1997 -----------------MSSKPTIVLVHGFWGGAAHWGNVIVELHGR--G
::: * . : *
TH_1085|M.thermoresistible__bu HWVLAICPTGVGERAHLLHAGVNLDTHITDVLAALHAHRVR--DAVLVGH
Mvan_3363|M.vanbaalenii_PYR-1 HHVLALTLTGVGERSHLMPGGVNLDTHIVDVLAAIDNDAATGADLILVGH
Mb0079c|M.bovis_AF2122/97 LRCIAPTWP-LGAHPKPLRPGADQTIGGVAGIVADVLAALELKDVVLVGN
Rv0077c|M.tuberculosis_H37Rv LRCIAPTWP-LGAHPKPLRPGADQTIGGVAGIVADVLAALELKDVVLVGN
MMAR_0223|M.marinum_M LRCFAPTWP-LGAHPRALGPDADRSITGVGDLVADVLATLDLHDVVLVGN
MUL_4891|M.ulcerans_Agy99 LRCFAPTWP-LGTHPRALGPDADRSITGVGDLVADVLATLDLHDVVLVGN
MSMEG_1797|M.smegmatis_MC2_155 HRAYAPTVAGHGHG-----ADTDVSIDDGVQSVIDYCRTRDLREIVLVGH
MAV_2604|M.avium_104 LRTLAVDLPGHGNKP---GDLATVTIADWVDSVIADIEEAGLGDIVIVGH
Mflv_2476|M.gilvum_PYR-GCK HDVTAVDLPGHGALAD-----QESTLANRRDAIVAAMQAGD-EKCVLVGH
MAB_0768|M.abscessus_ATCC_1997 FEDLHAVEN-------------PLTSLADDAARTRQMVQQIDGPVLLVGH
::**:
TH_1085|M.thermoresistible__bu SYGGMVITGVADRAG-PRVDSLVYL--DAFVPRDGESCWTLTTDDQRAWY
Mvan_3363|M.vanbaalenii_PYR-1 SYGGMVITGVADRIP-DRVDSLVFL--DAVVPRDGEACWDLVNDEERQWY
Mb0079c|M.bovis_AF2122/97 DTGGVVTQLVAVHYP-ERLGALVLTSCDAFEHFPPPILKPVILAAKSATL
Rv0077c|M.tuberculosis_H37Rv DTGGVVTQLVAVHYP-ERLGALVLTSCDAFEHFPPPILKPVILAAKSATL
MMAR_0223|M.marinum_M DTGGVVAQLVAVRHP-ERLGALVLTSCDAFEHFPPPILKPVILAAKSKKM
MUL_4891|M.ulcerans_Agy99 DTGGVVAQLVAVHHP-ERLGALVLTSCDAFEHFPPPILKPVILAAKSKKM
MSMEG_1797|M.smegmatis_MC2_155 SLGGTIIARVAEEIP-DRITRLIFWS--AFVPRPGRSITEEVEQPSTPAA
MAV_2604|M.avium_104 SMAGVTVPGVVAKLGSSRVREMVLAT--AFVPPQGSAIADTLGGPLAVFA
Mflv_2476|M.gilvum_PYR-GCK SGGGFDATLAADARP-DLVSHITYLA--AALPREGRTYPEAMAMRDDDNG
MAB_0768|M.abscessus_ATCC_1997 SYGGAVITEAGDLPN---VAGLVYVA--AFAPDAGESPGQLTEQLPPAAA
. .* . : : *
TH_1085|M.thermoresistible__bu GSVD-----------------ATGLAVPPLPFFDERATGHPLAALMQPIR
Mvan_3363|M.vanbaalenii_PYR-1 VKVD-----------------DSGFGVPPMPFFDDRATSHPLATVLQPLR
Mb0079c|M.bovis_AF2122/97 FRAA-----------------IQVMRAPAARNRAYAGLSHHNIDHLTRAW
Rv0077c|M.tuberculosis_H37Rv FRAA-----------------IQVMRAPAARNRAYAGLSHHNIDHLTRAW
MMAR_0223|M.marinum_M FRAA-----------------IQTMRSPAARRRAYGDLAHADIDELAQRW
MUL_4891|M.ulcerans_Agy99 FRAA-----------------IQTMRSPAARRRTYGDLAHADIDELAQRW
MSMEG_1797|M.smegmatis_MC2_155 EKQA-----------------PATGSSETLSLQVWRDVFVPDVDPEQAAT
MAV_2604|M.avium_104 RRAA-----------------RIGRPMKIPAPAARWAFCNGMTAAQRRLT
Mflv_2476|M.gilvum_PYR-GCK PVELGEDFDGDVGEMLGYLHFDDTGAMKFADFDGAWKYFYHDCDEETARW
MAB_0768|M.abscessus_ATCC_1997 ANLVP-----------------DSDGYLWIKQDKFRESFAQDLPEDAALV
TH_1085|M.thermoresistible__bu LSGTLES---------------IRRRVYVYATGWD-GESPLRPTYER---
Mvan_3363|M.vanbaalenii_PYR-1 LRGDLNG---------------FRRRIFVYALDWP-GESPLRPSYDR---
Mb0079c|M.bovis_AF2122/97 VRPALSNPAIA---------EDLRQLSLSLRTEVTTAVAARLPEFDKPAL
Rv0077c|M.tuberculosis_H37Rv VRPALSNPAIA---------EDLRQLSLSLRTEVTTAVAARLPEFDKPAL
MMAR_0223|M.marinum_M VQPALSNPAIA---------EDLRQLTLSLHTDVTRGVAARLPEFSKPAL
MUL_4891|M.ulcerans_Agy99 VQPALSNPAIA---------EDLRQLTLSLHTDVTRGVAARLPEFSKPAL
MSMEG_1797|M.smegmatis_MC2_155 WHALLSPEPRRPKTERLD-LRRFYRSTLPMHYIDAVDDRALPRGLDREAM
MAV_2604|M.avium_104 MSKLQAESARVPGEPVD---RSGLPADVPRTWILTTRDRALSVASQHASI
Mflv_2476|M.gilvum_PYR-GCK AFERLGPERFGDTTVTPVSVPNFWAADLPRSFIVCEQDQAMPRWLADT--
MAB_0768|M.abscessus_ATCC_1997 MAVTQKAPLASTFG---DAITAPAWRNKPSWYQVSTQDRMINPENERR-M
TH_1085|M.thermoresistible__bu LRRDPAWTTHALDAPHNLLREHTDAVLRILLDTAVPASGTKLQHVSV---
Mvan_3363|M.vanbaalenii_PYR-1 VRDDPTWICHELDGRHNLMRDRPADLLRILLSASQS--------------
Mb0079c|M.bovis_AF2122/97 IAWSADDVFFALENGQRLAATIPRARFEVIEGARTFSMVDSPDRLADQLS
Rv0077c|M.tuberculosis_H37Rv IAWSADDVFFALENGQRLAATIPRARFEVIEGARTFSMVDSPDRLADQLS
MMAR_0223|M.marinum_M IAWSADDKFFALEDGRRLAATIPNARLELIEGARTFSMLDRPDRLADLLS
MUL_4891|M.ulcerans_Agy99 IAWSADDKFFALEDGRRLAATIPNARLELIEGARTFSMLDRPDRLADLLS
MSMEG_1797|M.smegmatis_MC2_155 IERLKNVRVHRVRGGHEVLFTDPAGIAAVIVEAGVS--------------
MAV_2604|M.avium_104 AALGGVETVIPVDACHEVMFSHPERLAQILIERCRLRGVGIR--------
Mflv_2476|M.gilvum_PYR-GCK VARRLGVDQLTIDASHSPFLSRPREFADLLVHATTTTPVAPLRPL-----
MAB_0768|M.abscessus_ATCC_1997 AQRIKPRKTIELDASHASLASQPVAIADLIAEAAGDIA------------
: : . ::
TH_1085|M.thermoresistible__bu ------
Mvan_3363|M.vanbaalenii_PYR-1 ------
Mb0079c|M.bovis_AF2122/97 TVAVRT
Rv0077c|M.tuberculosis_H37Rv TVAVRT
MMAR_0223|M.marinum_M AVAVRT
MUL_4891|M.ulcerans_Agy99 AVAVRT
MSMEG_1797|M.smegmatis_MC2_155 ------
MAV_2604|M.avium_104 ------
Mflv_2476|M.gilvum_PYR-GCK ------
MAB_0768|M.abscessus_ATCC_1997 ------