For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ADVHNPNAPMLEQHTIGFVPASDRHGKVRDLFTLWFGTNIAPLPIVTGALGPSLFGLNLWWSLLAFVVGH VIGGALMALHSAQGPQMGIPQMIQSRGQFGTLGALIIVVIAAVMYLGFFASNIVLAGQSIHGIASAVPVN VGIIAGAVGSAIICIIGYRLIHFLNRIGTWVLGIAIVGGFIAILTSNLPADFLTRGGFNFAGWLATVSLG ALWQIAFAPYVSDYSRYLPKNVGVASTFWTTYLGCVLGSFLAFAFGAVVALAAPVGMDTMDGVKVTTGAL GTTMLVLFVLSVISHNALNLYGAVLSIITSVQTFRARWIPTAQARVFLSVVILVGASIVAIALSADFVSH FVDLVVALLVVLVPWTAINLIDFYIVHKGVYDLESIFRFDGGIYGRYNRAAVAAYILGIVVQIPFMATSL YTGPVAAHLGGADLSWLAGLIVTGPIYLLLVRRSPAPAIATTPQPETEVRNAESDLAAHSES*
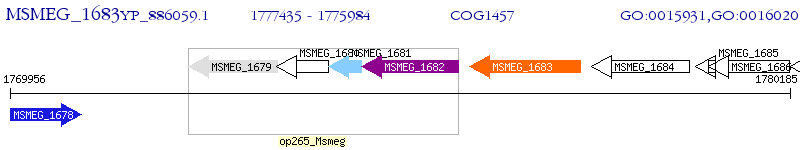
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1683 | - | - | 100% (483) | cytosine/purine/uracil/thiamine/allantoin permease family protein |
| M. smegmatis MC2 155 | MSMEG_1177 | - | 4e-97 | 41.43% (461) | cytosine/purines/uracil/thiamine/allantoin permease family |
| M. smegmatis MC2 155 | MSMEG_5653 | - | 1e-72 | 34.74% (475) | cytosine/purine/uracil/thiamine/allantoin permease family |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0055 | - | 6e-32 | 26.59% (455) | permease for cytosine/purines, uracil, thiamine, allantoin |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2025 | - | 4e-30 | 26.35% (482) | purine/cytosine permease |
| M. marinum M | MMAR_1212 | - | 9e-09 | 23.44% (448) | hypothetical protein MMAR_1212 |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2680 | - | 5e-06 | 22.89% (415) | hypothetical protein MUL_2680 |
| M. vanbaalenii PYR-1 | Mvan_3383 | - | 2e-34 | 27.19% (445) | permease for cytosine/purines, uracil, thiamine, allantoin |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1212|M.marinum_M MIDTTAPHSPVHSPSHGAGPANIKPDYHPRLTNDDLAPLR--EQTWGTYN
MUL_2680|M.ulcerans_Agy99 MIDTTAPHSPVHSPSHGAGPANIKPDYHPRLTNDDLAPLR--EQTWGTYN
MSMEG_1683|M.smegmatis_MC2_155 MADVHNPNAPMLEQHTIG-----------------FVPAS--DRHGKVRD
Mflv_0055|M.gilvum_PYR-GCK MHDLEAELT--------------------------VIPES--AKTTDLRG
Mvan_3383|M.vanbaalenii_PYR-1 MPQTSPGSTTGALTGEVFSGHQPSGVGDLSVETHGIAPISGDHRYGSPGR
MAB_2025|M.abscessus_ATCC_1997 MDEIQACCSATSLIASRESVGRDRVG---AIERAGIDYLPEAARGSSPKN
* : : . :
MMAR_1212|M.marinum_M IFAFWMSDVHSVGGYVTAGS-LFALGLASWQVLVSLVIAITIVYFLCNLV
MUL_2680|M.ulcerans_Agy99 IFAFWMSDVHSVGGYVTAGS-LFALGLASWQVLVSLVIAITIVYFLCNLV
MSMEG_1683|M.smegmatis_MC2_155 LFTLWFGTNIAPLPIVTGALGPSLFGLNLWWSLLAFVVGHVIGGALMALH
Mflv_0055|M.gilvum_PYR-GCK QFWIWAGANIAPINWILGAL-GVTMGLGLFETIAVLVVGNAIGMSLFGTF
Mvan_3383|M.vanbaalenii_PYR-1 LFTVWFAPQVNMSGVFTGTL-AIVLGLGFWLGLLAMVIGTVLGSLVVGYL
MAB_2025|M.abscessus_ATCC_1997 LFAVFVGGNLAFSTIIFGWL-PIVMGLTFWQAVSSSVTGLLIGLMLTAPM
* .: . . . :** : : * . : :
MMAR_1212|M.marinum_M ARPSQATGVPYPVICRTAFGVLGANIPAIIRGLIAVAWYGIQTYLASAAL
MUL_2680|M.ulcerans_Agy99 ARPSQATGVPYPVICRTAFGVLGANIPAIIRGLIAVAWYGIQTYLASAAL
MSMEG_1683|M.smegmatis_MC2_155 SAQGPQMGIPQMIQSRGQFGTLGALIIVVIAAVMYLGFFASNIVLAGQSI
Mflv_0055|M.gilvum_PYR-GCK VVIGQKTGVTGMVLSRMVFGRRGAYVPAAVQAALCIGWCAVNTWIVLDLV
Mvan_3383|M.vanbaalenii_PYR-1 STWGPRTGTGQLPNARMAFGG-TVVVPAALQWLSSIAWDALVGLFGGEAL
MAB_2025|M.abscessus_ATCC_1997 SLFGPRTGTNNPVSSGAHFGVRGRLIGSSLTLMFALVYAAIAVWTSGEAL
. * . ** : : : : . :
MMAR_1212|M.marinum_M DVVMIKL------FPGLARYAVVEDYGFLGLPALGWASYLILWVLQACVF
MUL_2680|M.ulcerans_Agy99 DVVMIKL------FPGLARYAVVEDYGFLGLPALGWASYLILWVLQACVF
MSMEG_1683|M.smegmatis_MC2_155 HGIASAV------PVNVG--IIAGAVGSAIICIIGYR--LIHFLNRIGTW
Mflv_0055|M.gilvum_PYR-GCK MALLGRIGLVDPELSNHAARIVVAGIIMTTQVVIAWFGYRLIAAFERWTV
Mvan_3383|M.vanbaalenii_PYR-1 SVLLGIP------------FWVAVLIVLGVQGVVGFFGYELIHRLQAVLT
MAB_2025|M.abscessus_ATCC_1997 IAGASRLFG---APQNHVAFLLGYGVIALEVILIALYGHATIVWAQKIII
: :. .
MMAR_1212|M.marinum_M WRGMEAIRKFIDFCGPAVYVVMFALSGYLISKAGWAAIDLNLGDVRYSGW
MUL_2680|M.ulcerans_Agy99 WRGMEAIRKFIDFCGPAVYVVIFALSGYLISTAGWAAIDLNLGDVRYSGW
MSMEG_1683|M.smegmatis_MC2_155 VLGIAIVGGFIAIL-----------------TSNLPADFLTRGGFNFAGW
Mflv_0055|M.gilvum_PYR-GCK PPTVAVLVAMTAVAWFG-----------LDVDWGYTGDAALTGAEHFTAI
Mvan_3383|M.vanbaalenii_PYR-1 VVLFVTFLVFTAKLVAG----------------HEIVVPAAVSGPDLVGA
MAB_2025|M.abscessus_ATCC_1997 PLAVVVLALGVLAYAPN--------------FHPMGSAQHGYALQEFWPT
. . .
MMAR_1212|M.marinum_M DAIPVMLGAIALVVSYFSGPMLNFGDFSRYG-RSFGAVRKGNFLGLPLN-
MUL_2680|M.ulcerans_Agy99 DAIPVMLGAIALVVSYFSGPMLNFGDFSRYG-RSFGAVRKGNFLGLPLN-
MSMEG_1683|M.smegmatis_MC2_155 LAT-VSLGALWQIA--FAPYVSDYSRYLPKN-VGVASTFWTTYLGCVLGS
Mflv_0055|M.gilvum_PYR-GCK TAVMTAIGIGWGIT--WLGYAGDYSRFVSSS-VPARKLFAASALGQFIP-
Mvan_3383|M.vanbaalenii_PYR-1 FVLEVTIAFSLAIS--WATYAADFSRYLPAD-VSPVRVFGFSTAGLALG-
MAB_2025|M.abscessus_ATCC_1997 WLFAVTVSVGGPLS--YAPSVGDYTRRISRSRYSDRQIAVAVSAGIFLGL
. :. : : :: . * :
MMAR_1212|M.marinum_M FLLFSLLVVVTASLTVPVFGELITDPVQTVARIDSTFAIILGASTFTIAT
MUL_2680|M.ulcerans_Agy99 FLLFSLLVVVTASLTVPVFGELITDPVQTVARIDSTFAIILGASTFTIAT
MSMEG_1683|M.smegmatis_MC2_155 FLAFAFGAVV--ALAAPVGMDTMDGVKVTTGALGTTMLVLFVLSVISHNA
Mflv_0055|M.gilvum_PYR-GCK --VVWLGALGATLATLSASSDPGEIIVDAYGVLAIPVLLLVVHGPLATNI
Mvan_3383|M.vanbaalenii_PYR-1 -YVFVQGVGIAAAGVVGEHTVEGVRSVMGGGLLGGLALLVIAVASIGSGV
MAB_2025|M.abscessus_ATCC_1997 FSTATFGAFTASTFSAPTASYVADMVATAPGWYVVPLLILAVLGGCGQGV
. . :: .
MMAR_1212|M.marinum_M IGIN-IVANFISPAFDFSN-VNPQRISWRTGGMIAAVGSALITPWNLYNH
MUL_2680|M.ulcerans_Agy99 IGIN-IVANFISPAFDFSN-VNPQRIGWRTGGMIAAVGSALITPWNLYNH
MSMEG_1683|M.smegmatis_MC2_155 LNLYGAVLSIITSVQTFRARWIPTAQARVFLSVVILVGASIVAIALSADF
Mflv_0055|M.gilvum_PYR-GCK LNIY-------TCAVSTQALDIHVNRRVLNIVIGVVAMAWVVYFVLNGDF
Mvan_3383|M.vanbaalenii_PYR-1 MNDY-------SGSLALQTLGVRVRR-PVSAVIVTVLAFALILWLHAADT
MAB_2025|M.abscessus_ATCC_1997 ISIY-------SSGLDLESIVPRLNRTQTTLIASGIAVVLMYLGVVVTDA
:. : : :
MMAR_1212|M.marinum_M PDVIHYTLETLGAFIGPLFGVLIADYYLVRKQHVVVDDLFS---LSHDGT
MUL_2680|M.ulcerans_Agy99 PDVIHYTLETLGAFIGPLFGVLIADYYLVRKQHVVVDDLFS---LSHDGT
MSMEG_1683|M.smegmatis_MC2_155 VSHFVDLVVALLVVLVPWTAINLIDFYIVHKGVYDLESIFR---FDGGIY
Mflv_0055|M.gilvum_PYR-GCK AATIDAWLVGIVGWLAPWAAVMLVHWFGVARRQVDPATLLTP--PNESTL
Mvan_3383|M.vanbaalenii_PYR-1 ATRFTDVLLLVSYWIPAFVAVVVVDWWRRAEGRPTVDPAAEV--TGRR--
MAB_2025|M.abscessus_ATCC_1997 IDSVTGMTVVLNSFAGSWVAINICGFFKANRGQYDPKALQRFGQDGRGGL
. : . .: : :: . .
MMAR_1212|M.marinum_M YWYRKGYNPAAVIATAVS--AAIAM--VPVLASDVYGMYTAAEYSWFIGC
MUL_2680|M.ulcerans_Agy99 YWYRKGYNPAAVIATAVS--AAIAM--APVLASDV---------------
MSMEG_1683|M.smegmatis_MC2_155 GRYNRAAVAAYILGIVVQ--IPFMA--TSLYTGPVAAHLGGADLSWLAGL
Mflv_0055|M.gilvum_PYR-GCK PAVRPSALISLAVGIVFT--WLFMYGMVPMMQGPIAVALGGVDLSWLAGG
Mvan_3383|M.vanbaalenii_PYR-1 --DAVVALVVFVVSYLAA--IPFMN--TSLIQGPVAMAWHGADIAYFVNL
MAB_2025|M.abscessus_ATCC_1997 YWFSHGFNVRAVVPFIAGSGLGLLTVQNDLYQAPFANIAGGIDVSLLLSV
: : : . .
MMAR_1212|M.marinum_M GVALVVYYLLATRG-RLAIAPPYHPVGAG-------------
MUL_2680|M.ulcerans_Agy99 ------------------------------------------
MSMEG_1683|M.smegmatis_MC2_155 IVTGPIYLLLVRRSPAPAIATTPQPETEVRNAESDLAAHSES
Mflv_0055|M.gilvum_PYR-GCK LAAGSLYAALEIPKARRA------------------------
Mvan_3383|M.vanbaalenii_PYR-1 AVAAVLYGGYRLMSRD--------------------------
MAB_2025|M.abscessus_ATCC_1997 VVGGGLYWLAQVSWPAPRVVSSGSSA----------------