For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ARTENDTWDLASSVGATATMVAAARALATNADEPAIDDPFAAPLVRAVGVDFFTKLVDDNFDLTALDPEA AEGLTRFANGMAARTRFFDDFFLDAARAGVRQFVILASGLDARAYRLPWPAGSVVFEIDQPDVIEFKTKA LADLGALPTTDRRAVAVDLRFDWPAALTEAGFDPAEPTAWIAEGLLGYLPPEAQDRLLDQIAELSAPGSR VAVEEIPAIEEEDHEEIAARMKDFSDRWRAHGFDLDFTELVFLGDRADVSSYLQGHGWKTTSMTSDELLV HNGLPRVDDDAQIGSVVYVTATR*
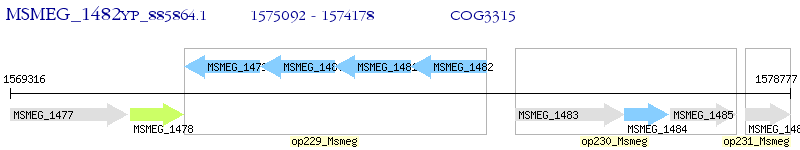
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1482 | - | - | 100% (304) | methyltransferase |
| M. smegmatis MC2 155 | MSMEG_1481 | - | 6e-92 | 59.03% (310) | methyltransferase |
| M. smegmatis MC2 155 | MSMEG_1479 | - | 2e-91 | 55.41% (305) | methyltransferase, putative, family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0752c | - | 1e-102 | 60.93% (302) | hypothetical protein Mb0752c |
| M. gilvum PYR-GCK | Mflv_5023 | - | 1e-112 | 64.82% (307) | putative methyltransferase |
| M. tuberculosis H37Rv | Rv0731c | - | 1e-102 | 60.93% (302) | hypothetical protein Rv0731c |
| M. leprae Br4923 | MLBr_02640 | - | 2e-66 | 45.69% (313) | hypothetical protein MLBr_02640 |
| M. abscessus ATCC 19977 | MAB_3787 | - | 1e-83 | 52.24% (312) | hypothetical protein MAB_3787 |
| M. marinum M | MMAR_1059 | - | 1e-101 | 61.76% (306) | O-methyltransferase |
| M. avium 104 | MAV_4441 | - | 1e-103 | 61.18% (304) | methyltransferase, putative, family protein |
| M. thermoresistible (build 8) | TH_0301 | - | 2e-95 | 59.42% (276) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0818 | - | 2e-99 | 60.13% (306) | O-methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_1346 | - | 1e-112 | 65.81% (310) | putative methyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0752c|M.bovis_AF2122/97 MTQTGSARFEGDSWDLASSVGLTATMVAAARAVAGRAPGALVNDQFAEPL
Rv0731c|M.tuberculosis_H37Rv MTQTGSARFEGDSWDLASSVGLTATMVAAARAVAGRAPGALVNDQFAEPL
MMAR_1059|M.marinum_M MTSTGSTRYDGDSWDLASSVGVTATMVAAARAMATRAENPLINDPYAEPL
MUL_0818|M.ulcerans_Agy99 MTSTGSTRYDGDSWDLASSVGVTATMVAAARAMATRAENPLINDPYAEPL
MAV_4441|M.avium_104 MTST---RYEGDTWDLASSVGVTATMVAAARAMATRADNPLINDPFAEPL
Mflv_5023|M.gilvum_PYR-GCK -----MPRTENDTWDLASSVGATATMVAAARAVATRAEDPVIDDPYAEPL
Mvan_1346|M.vanbaalenii_PYR-1 -----MARTDNDTWDLASSVGATATMVAAARAVATRAPRPVIDDPFAAPL
MSMEG_1482|M.smegmatis_MC2_155 -----MARTENDTWDLASSVGATATMVAAARALATNADEPAIDDPFAAPL
TH_0301|M.thermoresistible__bu -----MX-------------GG-----------ASRDPKELINDPFAAPL
MAB_3787|M.abscessus_ATCC_1997 -----MARTDDDSWDLASSVGATATMVAAQRALASHVPNPLINDPYAEPL
MLBr_02640|M.leprae_Br4923 ------MRTHDDTWDIKTSVGTTAVMVAAARAAETDRPDALIRDPYAKLL
* : * :* *
Mb0752c|M.bovis_AF2122/97 VRAVGVDFFVRMASGELDPDELA------EDEANGLRRFADAMAIRTHYF
Rv0731c|M.tuberculosis_H37Rv VRAVGVDFFVRMASGELDPDELA------EDEANGLRRFADAMAIRTHYF
MMAR_1059|M.marinum_M VRAVGVDLLTRLATGEFNVADLD---DDPQRPLGPLGDVADNMAVRTRFF
MUL_0818|M.ulcerans_Agy99 VRAVGVDLLTRLATGEFNVADLD---DDPQRPLGPIGDVADNMAVRTRFF
MAV_4441|M.avium_104 VKAVGVDLLSRLAGGELDPAELNDVHDGAAGSAGAMSRMADNMAVRTKFF
Mflv_5023|M.gilvum_PYR-GCK VRAVGIDFFAKLASGELTPEDLDVS---GDESPVGMSRFADGMAARTRFF
Mvan_1346|M.vanbaalenii_PYR-1 VQAVGVDFFTRLATGELTTEELDTN---DAESPVGMSRFADAMAARTRFF
MSMEG_1482|M.smegmatis_MC2_155 VRAVGVDFFTKLVDDNFDLTALDP------EAAEGLTRFANGMAARTRFF
TH_0301|M.thermoresistible__bu VRAVGIDFFTRIADGELTVADLDP------QSAPRVQANIDEVAVRTRFF
MAB_3787|M.abscessus_ATCC_1997 VRAVGIEYFTKLASGAFDPEQMAG----LVQLGITADGMANGMASRTWYY
MLBr_02640|M.leprae_Br4923 VTNTGAGALWEAMLDPSMVAKVEAI---DAEAAAMVEHMRSYQAVRTNFF
* .* : . . : . * ** ::
Mb0752c|M.bovis_AF2122/97 DNFFLDATRAG---IRQAVILASGLDSRAYRLRWPAGTIVFEVDQPQVID
Rv0731c|M.tuberculosis_H37Rv DNFFLDATRAG---IRQAVILASGLDSRAYRLRWPAGTIVFEVDQPQVID
MMAR_1059|M.marinum_M DDFFLDATRAG---LEQVVILASGLDARAYRLPWPPQTVVYEIDLPQVIE
MUL_0818|M.ulcerans_Agy99 DDFFLAATRAG---LEQVVILASGRDARAYRLPWPPQTVVYEIDLPQVID
MAV_4441|M.avium_104 DEFFLNATKAG---IAQVVILASGLDARAYRLAWPAGTVVYEVDQPQVID
Mflv_5023|M.gilvum_PYR-GCK DDFFTDACAAG---VRQAVILASGLDARGYRLTWPEGMTLFEIDQPEVIE
Mvan_1346|M.vanbaalenii_PYR-1 DDFFAEAMRAGPPPIRQGVILASGLDARAYRLSWPQDMVLFEIDQPEVLT
MSMEG_1482|M.smegmatis_MC2_155 DDFFLDAARAG---VRQFVILASGLDARAYRLPWPAGSVVFEIDQPDVIE
TH_0301|M.thermoresistible__bu DDHFAAATARG---IRQAVILAAGLDARAYRLRWPDGTVVYEIDQPRVIE
MAB_3787|M.abscessus_ATCC_1997 DTAFESSTAAG---VRQAVILASGLDTRAYRLTWPAGTTVYELDQPEVIT
MLBr_02640|M.leprae_Br4923 DTYFNNAVIDG---IRQFVILASGLDSRAYRLDWPTGTTVYEIDQPKVLA
* * : * : * ****:* *:*.*** ** ::*:* * *:
Mb0752c|M.bovis_AF2122/97 FKTTTLAGLGAAPTTDRRTVAVDLRDDWPTALQKAGFDNAQRTAWIAEGL
Rv0731c|M.tuberculosis_H37Rv FKTTTLAGLGAAPTTDRRTVAVDLRDDWPTALQKAGFDNAQRTAWIAEGL
MMAR_1059|M.marinum_M FKSRTLADLGAAPTADRRVVAVDLREDWPAALRAAGFDPNQPTAWSAEGL
MUL_0818|M.ulcerans_Agy99 FKSRTLADLGAAPTADRRVVAVDLREDWPAALRAAGFDPNQPTAWSAEGL
MAV_4441|M.avium_104 FKTTALAQLGAAPTAERRVVAVDLRDDWPAALRAAGFDPTRPTAWSAEGL
Mflv_5023|M.gilvum_PYR-GCK FKRTTLAGLGATPSTHLATVAIDLREDWPTALRDAGFDPSVPTAWIAEGL
Mvan_1346|M.vanbaalenii_PYR-1 FKAATLADLGVRPSTKLATVAVDLRNDWPAALAEAGFDASAPTAWIAEGL
MSMEG_1482|M.smegmatis_MC2_155 FKTKALADLGALPTTDRRAVAVDLRFDWPAALTEAGFDPAEPTAWIAEGL
TH_0301|M.thermoresistible__bu FKTRTLARLGAEPTAERRTVAADLRHDWPAALTAAGFDPAAPTVWIAEGL
MAB_3787|M.abscessus_ATCC_1997 FKTDTLAELGASPSAERRTVAIDLREDWPAALQAAGFDPEQPTVWSAEGL
MLBr_02640|M.leprae_Br4923 YKSTTLAEHGVTPTADRREVPIDLRQDWPPALRSAGFDPSARTAWLAEGL
:* :** *. *::. *. *** ***.** **** *.* ****
Mb0752c|M.bovis_AF2122/97 LGYLSAEAQDRLLDQITAQSVPGSQFATEVLRDINRLNEEELRGRMRRLA
Rv0731c|M.tuberculosis_H37Rv LGYLSAEAQDRLLDQITAQSVPGSQFATEVLRDINRLNEEELRGRMRRLA
MMAR_1059|M.marinum_M LGYLPPEAQDRLLDTVTELSAPGSRLAAECLSSVDPGEEEQIKERMQEVS
MUL_0818|M.ulcerans_Agy99 LGYLPPEAQDRLLDTVTELSAPGSRLAAECLSGVDPAEEEQIKERMQEVS
MAV_4441|M.avium_104 LGYLPPEAQDRLLDTITELSAPGSRLATESAPNPAPGEEEKLKERMQAIS
Mflv_5023|M.gilvum_PYR-GCK LGYLPPDAQDRLLDTIGELSASGSRLAVESVPSQQSADQDEMREKMKDST
Mvan_1346|M.vanbaalenii_PYR-1 LGYLPPDAQDRLLDTIGELSAPGSRLAVESVPTHHEADQEELREKMKESS
MSMEG_1482|M.smegmatis_MC2_155 LGYLPPEAQDRLLDQIAELSAPGSRVAVEEIPAIEEEDHEEIAARMKDFS
TH_0301|M.thermoresistible__bu FGYLPPDAQDRLLDQITDLSAVGSRLGVEGVPAHADADEDEVRQRMRILA
MAB_3787|M.abscessus_ATCC_1997 LIYLPPEAQDRLFASITELSAPGSRLVCEQVPGLETADFS----KARELT
MLBr_02640|M.leprae_Br4923 LMYLPATAQDGLFTEIGGLSAVGSRIAVETSPLHGDEWREQMQLRFRRVS
: **.. *** *: : *. **:. * . : : :
Mb0752c|M.bovis_AF2122/97 ERFRRHGLDLDMSGLVYFGD-RTDARTYLADHGWRTASASTTDLLAEHG-
Rv0731c|M.tuberculosis_H37Rv ERFRRHGLDLDMSGLVYFGD-RTDARTYLADHGWRTASASTTDLLAEHG-
MMAR_1059|M.marinum_M ARWRAHGFDVDMVGLVYFGD-RNEAVPYLSDRGWLLTSTPLPELRAANG-
MUL_0818|M.ulcerans_Agy99 ARWRAHGFDVDMVGLVYFGD-RNEAVPYLSDRGWLLTSTPLPELRAANG-
MAV_4441|M.avium_104 QRWRAHGFDLDMAGLVYFGE-RNEAAPYLAGHGWRLNSVTIRDLFAANG-
Mflv_5023|M.gilvum_PYR-GCK DRWRSHGFDLDFSELVFLGE-RANVPDYLRERGWAVTATPTNDLLTRYG-
Mvan_1346|M.vanbaalenii_PYR-1 DRWRSHGFDVDFSELVFLGE-RADVTDYLRDRGWTVDATPTNQLLTRYG-
MSMEG_1482|M.smegmatis_MC2_155 DRWRAHGFDLDFTELVFLGD-RADVSSYLQGHGWKTTSMTSDELLVHNG-
TH_0301|M.thermoresistible__bu ERWRAHGLDLDFSELIYLGD-RADVAGYLADRGWRCTALTANELLVRTG-
MAB_3787|M.abscessus_ATCC_1997 RQFAGETLDLDLESLVYTQE-RQMAADWLAEHGWTVITEENDELYARLN-
MLBr_02640|M.leprae_Br4923 DALG-FEQAVDVQELIYHDENRAVVADWLNRHGWRATAQSAPDEMRRVGR
:*. *:: : * . :* :** : : .
Mb0752c|M.bovis_AF2122/97 LPPID-GDDAPFGEVIYVSAELKQKHQDTR--
Rv0731c|M.tuberculosis_H37Rv LPPID-GDDAPFGEVIYVSAELKQKHQDTR--
MMAR_1059|M.marinum_M LAPAAVDDDGPSVDMLYVSGTLYTTPRPDPAP
MUL_0818|M.ulcerans_Agy99 LAPAAVDDDGPSVDMLYVSGTLYTTPRPDPAP
MAV_4441|M.avium_104 LDPLD-DDDTRMGEMLYTWG-IYE--------
Mflv_5023|M.gilvum_PYR-GCK LAPLD--TDEGFAEVVYVNAAR----------
Mvan_1346|M.vanbaalenii_PYR-1 LAPLD--SDEGFAEVVYVSATR----------
MSMEG_1482|M.smegmatis_MC2_155 LPRVD--DDAQIGSVVYVTATR----------
TH_0301|M.thermoresistible__bu LPPAE--NDGRVTEVVYLSSEKSPGKRAG---
MAB_3787|M.abscessus_ATCC_1997 LDPPNPLLRSIFPNIVYVDATLG---------
MLBr_02640|M.leprae_Br4923 WGDGVPMADDKDAFAEFVTAHRL---------
: .