For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVAPLTLQSKKRVHPVDEVPPVGKLATYGFQHVVAFYAGAVLVPILIATAIDLPTQDLVKLITADLFTCG IASIIQAVGFWKVGVRLPLLQGVTFAGVSPIIAIGLAHGGGAPSLLYVYGAVIVAGIFTFLIAPVFIKLL RFFPPVVTGTLITIIGLCLVPVGAHDAVTNPHTGEVDPTNLRWFLYALGTIAVIVAIQRIFSGFLATIAV LLGLVIGCVVAYALGDMNFDDVAQANMFGFTQPFLFGMPKFDFVACLTMIIVLMITAVESTGSTYATGEI VGKRIKASDIGNVLRADGLATTIGGVFNSFPYTAFSENVGLVRLTGVKSRWVVAAAGVIMILLGFLPKVA AVVSSIPQPVLGGAALTLFATVAVVGIQTLGKVDFTDHRNLIIVTTSLAAALWATSYPDVAQALPAGIDL LFGSGISIGAISAILLNVVFFHLGGRGARVAGSGTITLDEVNAMTEQRFKEVFGRVVQNVPWVLERVYTS RPFTDTNALRQAFQDALLAGSADQQLELVRAFPDLGAEDETGHALAVDHVALSNLDEDDHNDVVQLATAY REHFGFPLIICARETQHFDRILQNGWSRIDNSPAAEKSFALIEIAKIVKYRFDDLVADANPIAAARFGRL TELR
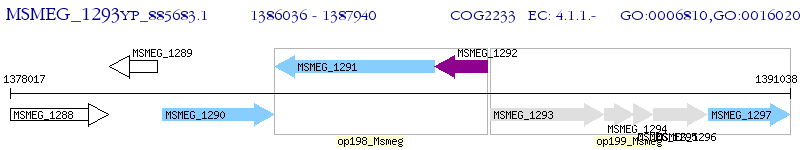
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1293 | - | - | 100% (634) | xanthine/uracil permeases family protein |
| M. smegmatis MC2 155 | MSMEG_2570 | - | 3e-92 | 40.93% (430) | xanthine/uracil permease |
| M. smegmatis MC2 155 | MSMEG_0810 | - | 5e-36 | 28.77% (438) | putative pyrimidine permease RutG |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0188 | - | 4e-35 | 28.31% (445) | uracil-xanthine permease |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2932c | - | 0.0 | 63.26% (645) | xanthine/uracil permease |
| M. marinum M | MMAR_4949 | - | 0.0 | 80.88% (633) | xanthine/uracil permease |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0456 | - | 6e-07 | 26.99% (163) | putative OHCU decarboxylase |
| M. vanbaalenii PYR-1 | Mvan_5275 | - | 0.0 | 80.57% (633) | uracil-xanthine permease |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1293|M.smegmatis_MC2_155 MVAPLTLQSKK-----RVHPVDEVPPVGKLATYGFQHVVAFYAGAVLVPI
MMAR_4949|M.marinum_M MSAPPTSPVR------RVHPVDEVLPIPKLATYGFQHVVAFYAGAVVVPI
Mvan_5275|M.vanbaalenii_PYR-1 MSAPTKSPKK------RVHPVDEVLPLPKLATYGFQHVVAFYAGAVLVPI
MAB_2932c|M.abscessus_ATCC_199 MSADTTRPAAPGAQRKKVHPVDEVPSTGRLVTLGIQHVIAFYAGAVLVPL
Mflv_0188|M.gilvum_PYR-GCK MTLTWKRVDSTTDP-DFVVAPDERLSWPSTLGLGAQHVVAMFGATFLVPV
MUL_0456|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_1293|M.smegmatis_MC2_155 LIATAIDLPTQDLVKLITADLFTCGIASIIQAVGFWKVGVRLPLLQGVTF
MMAR_4949|M.marinum_M IIANAIDLPHQDLVKLITADLFTCGVASIIQSVGFWKIGVRLPLLQGVAF
Mvan_5275|M.vanbaalenii_PYR-1 LIANAIGLSGEELVMLITADLFTCGIASIIQAVGFKNIGVRLPLLQGVTF
MAB_2932c|M.abscessus_ATCC_199 LIARAIDLDAEALTMLITADLFTCGVASILQAVGFWKIGVRLPLLQGVTF
Mflv_0188|M.gilvum_PYR-GCK LTG-----------FPPATTLLFSGVGTILFLL---ITGNRLPSYLGSSF
MUL_0456|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_1293|M.smegmatis_MC2_155 AGVSPIIAIGLAHGGGAPS--LLYVYGAVIVAGIFTFLIAPVFIKLLRFF
MMAR_4949|M.marinum_M AGVSPVIAIGLSHGGGTPS--LLYVYGAVIVAGLFTILIAPIFTKLLRFF
Mvan_5275|M.vanbaalenii_PYR-1 AGVAPIIAIGLAHGGGTES--LLYVYGAVIVAGVFTFLIAPMFVRLLKFF
MAB_2932c|M.abscessus_ATCC_199 ATLAPVIKIANDHGGGRVG--LLTVYGAVIAAGAFTFLIAPFFARLIRFF
Mflv_0188|M.gilvum_PYR-GCK SVIAPVTAATASHGTGSALGGIIAVGVLLIIIGAVVHVAGTRWIDIT--L
MUL_0456|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_1293|M.smegmatis_MC2_155 PPVVTGTLITIIGLCLVPVGAHDAVTNPHT--GEVDPTNLRWFLYALGTI
MMAR_4949|M.marinum_M PPVVTGTLITIIGLCLVPVGAGDAVTNPRT--GEHDPTNIRWLLYALGTI
Mvan_5275|M.vanbaalenii_PYR-1 PPVVTGTLITIIGLCLVPIGALDAVTNPAT--HEPDPTNLRWVLYALGTI
MAB_2932c|M.abscessus_ATCC_199 PPVVTGSVITIIGVSLLPVGIGDATRNPHSPVAAHDPGNGRWLMYALGTI
Mflv_0188|M.gilvum_PYR-GCK PPVVTGAIVALIGFNLAPAAKNNFEQGPLLG---------------FVTL
MUL_0456|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_1293|M.smegmatis_MC2_155 AVIVAIQRIFSGFLATIAVLLGLVIGCVVAYALGDMNFDDVAQANMFGFT
MMAR_4949|M.marinum_M AIIVAIQRVFSGFMATIAVLIGLVVGCVVAYGLGDMSFAKVDEASVIGFT
Mvan_5275|M.vanbaalenii_PYR-1 AVIVAIQRLFRGFMSTIAVLLGLVAGCTVAFTLGDMNFDRVAEASMFGFT
MAB_2932c|M.abscessus_ATCC_199 GAIVLMQRFFRGFWSTIAVLLGLVAGTVVAWLAGDTDFGRVGEADWIGFT
Mflv_0188|M.gilvum_PYR-GCK VLLVVVLAAFRGLLGRLAIFVAVAAGYLLAVFLGEVDTSGIAAAAWIGLP
MUL_0456|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_1293|M.smegmatis_MC2_155 QPFLFGMPKFDFVACLTMIIVLMITAVESTGSTYATGEIVGKRIKASDIG
MMAR_4949|M.marinum_M EPFVFGMPKFDFVACLTMIIVLMITSVESTGSTLATGEIVGKRIKAAQIG
Mvan_5275|M.vanbaalenii_PYR-1 PPFAFGMPRFDIVAIITMIIVMMIIAVESTGSTIATGEIVGKRVRPSDIG
MAB_2932c|M.abscessus_ATCC_199 SPFAFGAPRFDVIAVVSLIVVLMVTAVESTGAVFATAEIVGKRVRGADIA
Mflv_0188|M.gilvum_PYR-GCK D---FQAPTFTLSVLPLFLPAVVALVAENIGHVKSVGQMT-KRDMDPLTG
MUL_0456|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_1293|M.smegmatis_MC2_155 NVLRADGLATTIGGVFNSFPYTAFSENVGLVRLTGVKSRWVVAAAGVIMI
MMAR_4949|M.marinum_M NVLRADGLATVIGGIFNSFPYTAFSENVGLVRLTRVKSRWVVAAAGVVMI
Mvan_5275|M.vanbaalenii_PYR-1 NVLRADGVATTIGGIFNSFPYTAFSENVGLVRLTGVKSRWVVAAAGVIMI
MAB_2932c|M.abscessus_ATCC_199 ATVRADGIATVIGGTFNSFPYTAFSENVGLVRLTGVKSRWVVAAAGVIMM
Mflv_0188|M.gilvum_PYR-GCK RALAADGVATTLAGFGGGSATTTYAENIGVMAATRVYSTAPYWIAALVAI
MUL_0456|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_1293|M.smegmatis_MC2_155 LLGFLPKVAAVVSSIPQPVLGGAALTLFATVAVVGIQTLGKVDFTDHRNL
MMAR_4949|M.marinum_M VLGFLPKVAAVVSAVPSPVLGGAALTLFATVAVVGIQTLGKVDFTDHRNL
Mvan_5275|M.vanbaalenii_PYR-1 LLGFLPKVAAIVASIPNPVLGGAALTLFATVAVVGIQTLGKVDFTDHRNV
MAB_2932c|M.abscessus_ATCC_199 VLGVLPKTAKIVESIPAPVLGGAAMTLFATVAVVGIQTLGKVDFNDHRNL
Mflv_0188|M.gilvum_PYR-GCK LLSLCPKVGAVIAAIPAGVLGGATVVLYGLVGIIGIRIWLTNHVDFSKPV
MUL_0456|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_1293|M.smegmatis_MC2_155 IIVTTSLAAALWATS--------------------YPDVAQALPAGIDLL
MMAR_4949|M.marinum_M IIVTTSLALALWVTS--------------------YPDVAESMPSGVDLI
Mvan_5275|M.vanbaalenii_PYR-1 IIVTTSLALALLVTS--------------------YPDISSQLPAGLDIL
MAB_2932c|M.abscessus_ATCC_199 IIASTSLAMGMYVQFSQSSGPATVLDHGRGIAVPALPGIDQAVPGLLQIP
Mflv_0188|M.gilvum_PYR-GCK NQMTAAIPLIIGIAD---------------------------FTWSAGSL
MUL_0456|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_1293|M.smegmatis_MC2_155 FGSGISIGAISAILLNVVFFHLGGRGARVAGSGTITLDEVNAMTEQRFKE
MMAR_4949|M.marinum_M FGSGISIGAVAAIVLNIVFFHTGSHGPRVAGGGSITLHEVNAMTRERFTE
Mvan_5275|M.vanbaalenii_PYR-1 FGSGISTGAITAIVLNIVFFHIVKKGPRVAGSGSITLDEVNAMTRERFTE
MAB_2932c|M.abscessus_ATCC_199 FSTGITMGAITAIVLNLLFFHTGSRGVAVAGGGAIRLAQVNEMTLEQFRR
Mflv_0188|M.gilvum_PYR-GCK TFTGIALGSIAALLIYHGMRLLGFR-PGRERPSTVYADPAGPAPEPR---
MUL_0456|M.ulcerans_Agy99 -----------------------------MTKPALGLDGFNRLTDRQRMH
:: . . :
MSMEG_1293|M.smegmatis_MC2_155 VFGRVVQNVPWVLERVYTSRPFTDTNALRQAFQDALLAGSADQQLELVRA
MMAR_4949|M.marinum_M VFGHVVQDVPWVIDRAYEHRPFADTAALREAFQDAVLTGSSEQQMELLNA
Mvan_5275|M.vanbaalenii_PYR-1 VFGHVVQGVSWAADRAFDQRPFADTAALREAFQDAVLTGNAEQQQELLDA
MAB_2932c|M.abscessus_ATCC_199 TFGGLVQDVHWVVDRAYRQRPFADAHDLRSAFQEAMLTGSDEEQLELIGS
Mflv_0188|M.gilvum_PYR-GCK --------------------------------------------------
MUL_0456|M.ulcerans_Agy99 LLFGVCSTTIWAR-RVLAGAPFDDVGALLDRADRVLAQLPEAEIDAALDG
MSMEG_1293|M.smegmatis_MC2_155 FPDLGAEDETGHALAVDHVALSNLDEDDHNDVVQLATAYREHFGFPLIIC
MMAR_4949|M.marinum_M FHDLGAEDETGRALAVDHVSLSNLDERDHNVVVELASAYREHFGFPLIIC
Mvan_5275|M.vanbaalenii_PYR-1 FHDLGAEDETGQVLAVDHVALSNLDEEDHQDVVTLAAAYREHFGFPLIIC
MAB_2932c|M.abscessus_ATCC_199 FPDLGAEDEAGEMVAVDHTILQTLDEAELNNVVSLATAYREHFGFPLVVC
Mflv_0188|M.gilvum_PYR-GCK --------------------------------------------------
MUL_0456|M.ulcerans_Agy99 HPRIGARADN-ASSAREQALVAGAGEAVRAELAEKNRKYEEKFGYVYLVC
MSMEG_1293|M.smegmatis_MC2_155 ARETQHFDRILQNGWSRIDNSPAAEKSFALIEIAKIVKYRFDDLVADANP
MMAR_4949|M.marinum_M ARETERFDRVLKNGWSRMDNSPATERAYALIEAAKIANYRFSDLVADANP
Mvan_5275|M.vanbaalenii_PYR-1 ARETERFDRVLRNGWSRMDNSPATEKAYALIEVAKIANYRFTDLVADANP
MAB_2932c|M.abscessus_ATCC_199 ARETERYERLLRSGWSRMENSATSERSFALIEIAKIANYRFDDLVADANP
Mflv_0188|M.gilvum_PYR-GCK --------------------------------------------------
MUL_0456|M.ulcerans_Agy99 ATG-RSAEDLLETLTDRLDNDPETERRVMRSELAKINRLRLQRLLREPA-
MSMEG_1293|M.smegmatis_MC2_155 IAAARFGRLTELR---
MMAR_4949|M.marinum_M IASARLSRLNELH---
Mvan_5275|M.vanbaalenii_PYR-1 IAAARFGRLNELS---
MAB_2932c|M.abscessus_ATCC_199 ITAARFGRFEDFHATR
Mflv_0188|M.gilvum_PYR-GCK ----------------
MUL_0456|M.ulcerans_Agy99 ----------------