For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNGLMQPGVWGVLATPFSGSTLEVDDNGVAVLAEHAQAVGATGLTVLGVFGEAAKLSWEERRRVLETVLD TTKLPLVVGCTSLATAPVIDEARMAVELVGDRLAGVMVQANSADRTTLATHLRAVNEATGAHVVVQDYPV ISGVHVPAGVLGDVVAQAPFVTAVKAEAPPTPVAVAELTRRVDVPVFGGLGGINLLDELACGSAGAMTGF SYPEALVATVRAWRDGDRMKARAALLDHLPLITFEQQVGIALAVRKECLRRRGLIAESGVRPPGRSLPDC LAPVLATHIDTVEGR
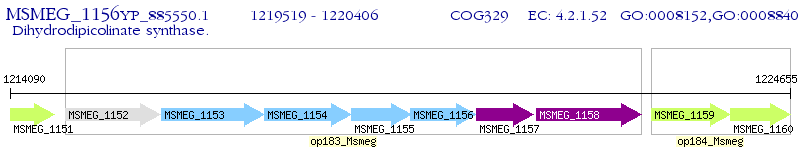
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1156 | - | - | 100% (295) | dihydrodipicolinate synthetase |
| M. smegmatis MC2 155 | MSMEG_2684 | dapA | 2e-09 | 26.36% (258) | dihydrodipicolinate synthase |
| M. smegmatis MC2 155 | MSMEG_0877 | - | 7e-06 | 24.92% (305) | dihydrodipicolinate synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2774c | dapA | 6e-08 | 25.39% (256) | dihydrodipicolinate synthase |
| M. gilvum PYR-GCK | Mflv_0676 | - | 5e-08 | 30.77% (169) | dihydrodipicolinate synthetase |
| M. tuberculosis H37Rv | Rv2753c | dapA | 6e-08 | 25.39% (256) | dihydrodipicolinate synthase |
| M. leprae Br4923 | MLBr_01513 | dapA | 5e-08 | 25.48% (259) | dihydrodipicolinate synthase |
| M. abscessus ATCC 19977 | MAB_1047c | - | 3e-09 | 26.53% (294) | dihydrodipicolinate synthetase DapA |
| M. marinum M | MMAR_2154 | dapA_1 | 1e-12 | 26.85% (257) | dihydrodipicolinate synthase DapA_1 |
| M. avium 104 | MAV_3450 | - | 3e-13 | 26.05% (261) | dihydrodipicolinate synthetase family protein |
| M. thermoresistible (build 8) | TH_0834 | dapA | 9e-07 | 23.44% (256) | PROBABLE DIHYDRODIPICOLINATE SYNTHASE DAPA (DHDPS) |
| M. ulcerans Agy99 | MUL_1739 | dapA_1 | 7e-12 | 26.83% (246) | dihydrodipicolinate synthase DapA_1 |
| M. vanbaalenii PYR-1 | Mvan_0452 | - | 1e-15 | 27.49% (291) | dihydrodipicolinate synthetase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2154|M.marinum_M -----MAEPGAPTIHGIIAYPVTPFT---AEGD-VDARRLAALVDRLVCS
MUL_1739|M.ulcerans_Agy99 -----MAEPGAPTIHGIIAYPVTPFT---AEGD-VDARRLAALVDRLVCS
MAV_3450|M.avium_104 -----MTEP---KIHGIIAYPVTPFA---DNG--IDTQRLAALVERLVSA
Mvan_0452|M.vanbaalenii_PYR-1 -----MTTATT-EIHGIIAYPVTPFT---SNGEDIDTGKLAALVEQLVAT
MAB_1047c|M.abscessus_ATCC_199 -----MTSTP--QFHGIIAYPVTPFL---PDDT-VDIDRLAELVSRLVDD
Mb2774c|M.bovis_AF2122/97 --MTTVGFDVAARLGTLLTAMVTPFS---GDGS-LDTATAARLANHLVDQ
Rv2753c|M.tuberculosis_H37Rv --MTTVGFDVAARLGTLLTAMVTPFS---GDGS-LDTATAARLANHLVDQ
MLBr_01513|M.leprae_Br4923 --MTTVGFDVPARLGTLLTAMVTPFD---ADGS-VDTAAATRLANRLVDA
TH_0834|M.thermoresistible__bu --VSTGGFDVTAQLGTVLTAMVTPFK---KDGS-LDLDAAASLANRLVDA
MSMEG_1156|M.smegmatis_MC2_155 --------MNGLMQPGVWGVLATPFS---GSTLEVDDNGVAVLAEHAQAV
Mflv_0676|M.gilvum_PYR-GCK MRDTKLTVDDITGVVGIIPTPSIPTADQPGTAFSVDLDEAAKLADAMVRG
: * :* : *..
MMAR_2154|M.marinum_M GVHAIAALGSTGELAYLDEAEFDTAVDTAVAAVSGRLPVVVGVSDVTTAK
MUL_1739|M.ulcerans_Agy99 GVHAIAALGSTGELAYLDEAEFDTAVDTAVAAVSGRLPVVVGVSDVTTAK
MAV_3450|M.avium_104 GVHAIAPLGSTGELAYLEEPEVDAVIDTTLSAVAGRVPVIVGVSDVTTAK
Mvan_0452|M.vanbaalenii_PYR-1 GAHAIAPLGSTGESAYLTEAEFDTVVDTTVGVVNGRVPVIVGASDLTTAN
MAB_1047c|M.abscessus_ATCC_199 GAHAIAPLGSTGESAYLEEREFDAVVDTTVAAVNKRVPVIVGASDLTTAN
Mb2774c|M.bovis_AF2122/97 GCDGLVVSGTTGESPTTTDGEKIELLRAVLEAVGDRARVIAGAGTYDTAH
Rv2753c|M.tuberculosis_H37Rv GCDGLVVSGTTGESPTTTDGEKIELLRAVLEAVGDRARVIAGAGTYDTAH
MLBr_01513|M.leprae_Br4923 GCDGLVLSGTTGESPTTTDDEKLQLLRVVLEAVGDRARVIAGAGSYDTAH
TH_0834|M.thermoresistible__bu GCDGLVVSGTTGEAPTTTDDEKLQLLRTVLDAVGDRARIIAGAGTYDTAH
MSMEG_1156|M.smegmatis_MC2_155 GATGLTVLGVFGEAAKLSWEERRRVLETVLDTT--KLPLVVGCTSLATAP
Mflv_0676|M.gilvum_PYR-GCK GVDVLMTTGTFGECASLTWDELQSFVATVVDAVAGRIPVFAGATTLNTRD
* : * ** . * : ..: .. : :..* *
MMAR_2154|M.marinum_M TIRRARYAQHVGADAVMILPVSYWKLNEHEIFQHYRSVGDAIT-IPIMAY
MUL_1739|M.ulcerans_Agy99 TIRRARYAQHVGADAVMILPVSYWKLNEHEIFQHYRSVGDAIT-IPIMAY
MAV_3450|M.avium_104 TIRRAQYAQRAGADAVMILPVSYWKLTEREIFHHYRSISDAIS-IPIMAY
Mvan_0452|M.vanbaalenii_PYR-1 TIRRGKYAQRAGADALMILPVSYWKLTDREIARHYASVAAATD-LPIMAY
MAB_1047c|M.abscessus_ATCC_199 TIRRARHAQQAGADAVMVLPISYWKLSDREIAQHYASVAAAID-IPLMAY
Mb2774c|M.bovis_AF2122/97 SIRLAKACAAEGAHGLLVVTPYYSKPPQRGLQAHFTAVADATE-LPMLLY
Rv2753c|M.tuberculosis_H37Rv SIRLAKACAAEGAHGLLVVTPYYSKPPQRGLQAHFTAVADATE-LPMLLY
MLBr_01513|M.leprae_Br4923 SVRLVKACAGEGAHGLLVVTPYYSKPPQTGLFAHFTAVADATE-LPVLLY
TH_0834|M.thermoresistible__bu SVRLAKACQDAGAHGLLLVTPYYSRPPQDGLYAHFTTIADAVE-VPIVLY
MSMEG_1156|M.smegmatis_MC2_155 VIDEARMAVELVGDRLAGVMVQANSADRTTLATHLRAVNEATG-AHVVVQ
Mflv_0676|M.gilvum_PYR-GCK TIARGRRLGELGADGLFVGRPMWLPLDDAGIVRFYRDVAEAVPNMALVVY
: : .. : : . : * ::
MMAR_2154|M.marinum_M NNPATSGVDMRPELLVRMFESIDNLTMVKESTGDLSRMRRIAELSGGRLP
MUL_1739|M.ulcerans_Agy99 NNPATSGVDMRPELLVRMFESIDNLTMVKESTGDLSRMRRIAELSGGRLP
MAV_3450|M.avium_104 NNPATSGVDMPPELLVRMFESIDNVAMVKESTGDLTRMRRIAELSGNRLP
Mvan_0452|M.vanbaalenii_PYR-1 NNPATSGIDMSPELLVRMFTDIDNVAMVKESTGDLSRMQRIKELTGGALP
MAB_1047c|M.abscessus_ATCC_199 NNPATSGVDMKPELLVSMFGDIDNFTMVKESTGDLNRMLEIKRLTNGLLP
Mb2774c|M.bovis_AF2122/97 DIPGRSAVPIEPDTIR-ALASHPNIVGVKDAKADLHSGAQIMADTG--LA
Rv2753c|M.tuberculosis_H37Rv DIPGRSAVPIEPDTIR-ALASHPNIVGVKDAKADLHSGAQIMADTG--LA
MLBr_01513|M.leprae_Br4923 DIPGRSVVPIEPDTIR-ALASHPNIVGVKEAKADLYSGARIMADTG--LA
TH_0834|M.thermoresistible__bu DIPPRSVVPLAWDTIR-RLSEHRNIVAIKDAKGDLHGAAQIIAETG--LA
MSMEG_1156|M.smegmatis_MC2_155 DYPVISGVHVPAGVLGDVVAQAPFVTAVKAEAPPTPVAVAELTRRVDVPV
Mflv_0676|M.gilvum_PYR-GCK DNPGAFKGKIGTPAYEALSQIPQVVAAKHLGLLSGSAFLSDLRAVGGRVR
: * : .. :
MMAR_2154|M.marinum_M FYNGSN--PLVLDALKVGAAGWCTAAPNLWPQPCIDLYEAVCAGDLEKAQ
MUL_1739|M.ulcerans_Agy99 FYNGSN--PLVLDALKVGAAGWCTAAPNLWPQPCIDLYEAVCAGDLEKAQ
MAV_3450|M.avium_104 FYNGSN--PLVLDALKAGASGWCTAAPCLRPQPCIDLYDAVRANDLDTAQ
Mvan_0452|M.vanbaalenii_PYR-1 FYNGSN--PLVLDALTEGAAGWCTAAPNLRPQPCLDLYTAVRDGERSRAQ
MAB_1047c|M.abscessus_ATCC_199 FYNGSN--PLVLDALNAGASGWCTAAPCLAPQPCLDLYEAVRAGRDDDAA
Mb2774c|M.bovis_AF2122/97 YYSGDD--ALNLPWLAMGATGFISVIAHLAAGQLRELLSAFGSGDIATAR
Rv2753c|M.tuberculosis_H37Rv YYSGDD--ALNLPWLAMGATGFISVIAHLAAGQLRELLSAFGSGDIATAR
MLBr_01513|M.leprae_Br4923 YYSGDD--ALNLPWLAVGAIGFISVISHLAAGQLRELLSAFGSGDITTAR
TH_0834|M.thermoresistible__bu YYSGDD--SLNLPWLAVGAVGFISVWGHLAAGQLRDMLTAFNSGDMTTAR
MSMEG_1156|M.smegmatis_MC2_155 FGGLGG--INLLDELACGSAGAMTGFS--YPEALVATVRAWRDGDRMKAR
Mflv_0676|M.gilvum_PYR-GCK LLPLETDWYYFARLFPEEVTACWSGNVACGPAPVTHLRDLIRARRWDDCQ
: . : . .
MMAR_2154|M.marinum_M ILYDDLKPLLEFIVAGG-------LATTV-KAGLGLLGFPVGD-PRPPLL
MUL_1739|M.ulcerans_Agy99 ILYDDLKPLLEFIVAGR-------LATTV-KAGLGLLGFPVGD-PRPPLL
MAV_3450|M.avium_104 RLYDDLKPLLTFIVAGG-------LATTV-KAGLELLDFKVGD-PRAPLL
Mvan_0452|M.vanbaalenii_PYR-1 EIYDDLAPLLRFIVAGG-------LPTTV-KAGLELLGVSVGD-PRAPLL
MAB_1047c|M.abscessus_ATCC_199 SLYTALKPLLQFIVAGG-------LPTTV-KAGLELQGRPVGD-PRRPLL
Mb2774c|M.bovis_AF2122/97 KINIAVAPLCNAMSRLG-------GVTLS-KAGLRLQGIDVGD-PRLPQV
Rv2753c|M.tuberculosis_H37Rv KINIAVAPLCNAMSRLG-------GVTLS-KAGLRLQGIDVGD-PRLPQV
MLBr_01513|M.leprae_Br4923 KINVAIGPLCSAMDRLG-------GVTMS-KAGLRLQGIDVGD-PRLPQM
TH_0834|M.thermoresistible__bu KIAVTLSPLNAAQTRLG-------GVSMS-KAGLRLLGIEVGD-PRLPQV
MSMEG_1156|M.smegmatis_MC2_155 AALLDHLPLITFEQQVG-------IALAVRKECLRRRGLIAESGVRPPGR
Mflv_0676|M.gilvum_PYR-GCK ALTDELEGALETLYPGGNFAEFLKYSIQIDNAQFQAAGFMRTGPTRPPYT
: : . . * *
MMAR_2154|M.marinum_M PLDEHGRA----------KLGRLLAGC-----
MUL_1739|M.ulcerans_Agy99 PLDEHGRA----------KLGRLLAGC-----
MAV_3450|M.avium_104 PLDEQGRA----------ELRKLLAA------
Mvan_0452|M.vanbaalenii_PYR-1 ALDDTGRA----------ELKTLLAVA-----
MAB_1047c|M.abscessus_ATCC_199 PLDPEGRE----------TLKGILASTP----
Mb2774c|M.bovis_AF2122/97 AATPEQID----------ALAADMRAASVLR-
Rv2753c|M.tuberculosis_H37Rv AATPEQID----------ALAADMRAASVLR-
MLBr_01513|M.leprae_Br4923 PATAEQID----------ELAVDMRAASVLR-
TH_0834|M.thermoresistible__bu PPGPEQLR----------LLADDMRAAAVLR-
MSMEG_1156|M.smegmatis_MC2_155 SLPDCLAP----------VLATHIDTVEGR--
Mflv_0676|M.gilvum_PYR-GCK EVPESYLAGGREAGKNWAALQQRYASLAVSNH
*