For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTRELTSDLAGIIPPLTTPLTADGDVDTASLERLCGFLLDAGVDGLFVCGSSGEVALLSDAQRAKAIQVA AGAASGQVPVLGGVIDTGTARVVDNALAAQKAGADAVVATPPFYVAPHADEVIRHYRLIAAAVDVPVVAY DIPSATHSPLTRSVVAQLAGEKTVAALKDSSGDLDGFRAHLAANADTGLRVFTGSETMTDVALNLGAHGA VPGLANVDPHGYVRIRQAARVGDWATAASEQERLRRIFAIVDVADRTRIGLTAGALGAFKAAQYLRGVID TPRCADPLLPLESSEIDAVRTILEREGVLG
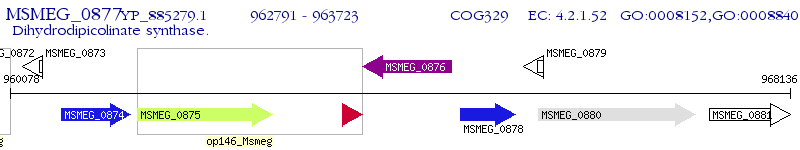
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0877 | - | - | 100% (310) | dihydrodipicolinate synthase |
| M. smegmatis MC2 155 | MSMEG_2684 | dapA | 1e-27 | 27.78% (306) | dihydrodipicolinate synthase |
| M. smegmatis MC2 155 | MSMEG_6133 | - | 1e-14 | 29.34% (167) | 5-dehydro-4-deoxyglucarate dehydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2774c | dapA | 1e-26 | 27.18% (309) | dihydrodipicolinate synthase |
| M. gilvum PYR-GCK | Mflv_4000 | - | 2e-24 | 28.34% (314) | dihydrodipicolinate synthase |
| M. tuberculosis H37Rv | Rv2753c | dapA | 1e-26 | 27.18% (309) | dihydrodipicolinate synthase |
| M. leprae Br4923 | MLBr_01513 | dapA | 4e-27 | 26.62% (308) | dihydrodipicolinate synthase |
| M. abscessus ATCC 19977 | MAB_3084c | - | 5e-30 | 28.81% (302) | dihydrodipicolinate synthase |
| M. marinum M | MMAR_2154 | dapA_1 | 3e-29 | 30.99% (284) | dihydrodipicolinate synthase DapA_1 |
| M. avium 104 | MAV_3644 | dapA | 3e-29 | 29.47% (302) | dihydrodipicolinate synthase |
| M. thermoresistible (build 8) | TH_0834 | dapA | 5e-29 | 29.27% (287) | PROBABLE DIHYDRODIPICOLINATE SYNTHASE DAPA (DHDPS) |
| M. ulcerans Agy99 | MUL_1739 | dapA_1 | 1e-29 | 32.16% (283) | dihydrodipicolinate synthase DapA_1 |
| M. vanbaalenii PYR-1 | Mvan_0452 | - | 2e-28 | 30.56% (301) | dihydrodipicolinate synthetase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2774c|M.bovis_AF2122/97 --MTTVGFDVAARLGTLLTAMVTPFSGDG-SLDTATAARLANHLVDQGCD
Rv2753c|M.tuberculosis_H37Rv --MTTVGFDVAARLGTLLTAMVTPFSGDG-SLDTATAARLANHLVDQGCD
MAV_3644|M.avium_104 --MSTVGFDAPARLGTVLTAMVTPFAADG-SLDTAAAARLANHLVDSGCD
MLBr_01513|M.leprae_Br4923 --MTTVGFDVPARLGTLLTAMVTPFDADG-SVDTAAATRLANRLVDAGCD
Mflv_4000|M.gilvum_PYR-GCK MPVSTSGFDAPARLGTLLTAMATPFKPDG-SLDIETAARLATRLVDAGCD
TH_0834|M.thermoresistible__bu --VSTGGFDVTAQLGTVLTAMVTPFKKDG-SLDLDAAASLANRLVDAGCD
MAB_3084c|M.abscessus_ATCC_199 ----------MARLGTVLTAMVTPFDAEG-ALDVDTAVRLAAHLVDAGCD
MMAR_2154|M.marinum_M -----MAEPGAPTIHGIIAYPVTPFTAEG-DVDARRLAALVDRLVCSGVH
MUL_1739|M.ulcerans_Agy99 -----MAEPGAPTIHGIIAYPVTPFTAEG-DVDARRLAALVDRLVCSGVH
Mvan_0452|M.vanbaalenii_PYR-1 ------MTTATTEIHGIIAYPVTPFTSNGEDIDTGKLAALVEQLVATGAH
MSMEG_0877|M.smegmatis_MC2_155 -----MTRELTSDLAGIIPPLTTPLTADG-DVDTASLERLCGFLLDAGVD
. : ::. .**: :* :* * *: * .
Mb2774c|M.bovis_AF2122/97 GLVVSGTTGESPTTTDGEKIELLRAVLEAVGDRARVIAGAGTYDTAHSIR
Rv2753c|M.tuberculosis_H37Rv GLVVSGTTGESPTTTDGEKIELLRAVLEAVGDRARVIAGAGTYDTAHSIR
MAV_3644|M.avium_104 GLVVSGTTGESPTTTDDEKRELLRVVLEAVGDRARVIAGAGTYDTAHSVR
MLBr_01513|M.leprae_Br4923 GLVLSGTTGESPTTTDDEKLQLLRVVLEAVGDRARVIAGAGSYDTAHSVR
Mflv_4000|M.gilvum_PYR-GCK GLVVSGTTGESPTTTDDEKILLLRTVLEAVGDRARIVAGAGTYDTAHSIH
TH_0834|M.thermoresistible__bu GLVVSGTTGEAPTTTDDEKLQLLRTVLDAVGDRARIIAGAGTYDTAHSVR
MAB_3084c|M.abscessus_ATCC_199 GLVLSGTTGESPVTSDAEKLTLLREVVAAVGDRARIIAGSGTYDTAHSIK
MMAR_2154|M.marinum_M AIAALGSTGELAYLDEAEFDTAVDTAVAAVSGRLPVVVGVSDVTTAKTIR
MUL_1739|M.ulcerans_Agy99 AIAALGSTGELAYLDEAEFDTAVDTAVAAVSGRLPVVVGVSDVTTAKTIR
Mvan_0452|M.vanbaalenii_PYR-1 AIAPLGSTGESAYLTEAEFDTVVDTTVGVVNGRVPVIVGASDLTTANTIR
MSMEG_0877|M.smegmatis_MC2_155 GLFVCGSSGEVALLSDAQRAKAIQVAAGAASGQVPVLGGVIDTGTARVVD
.: *::** . : : : . ....: :: * **. :
Mb2774c|M.bovis_AF2122/97 LAKACAAEGAHGLLVVTPYYSKPPQRGLQAHFTAVADATELPMLLYDIPG
Rv2753c|M.tuberculosis_H37Rv LAKACAAEGAHGLLVVTPYYSKPPQRGLQAHFTAVADATELPMLLYDIPG
MAV_3644|M.avium_104 LAKACAAEGAHGLLVVTPYYSKPPQRGLIAHFTAVADATELPVLLYDIPG
MLBr_01513|M.leprae_Br4923 LVKACAGEGAHGLLVVTPYYSKPPQTGLFAHFTAVADATELPVLLYDIPG
Mflv_4000|M.gilvum_PYR-GCK LAKASAAEGAHGLLVVTPYYSRPPQAGLVAHFTAVADATDLPNILYDIPP
TH_0834|M.thermoresistible__bu LAKACQDAGAHGLLLVTPYYSRPPQDGLYAHFTTIADAVEVPIVLYDIPP
MAB_3084c|M.abscessus_ATCC_199 LSRECEAAGAHGLLVVTPYYSRPSQAGLLAHFTAVADAVNVPVILYDIPP
MMAR_2154|M.marinum_M RARYAQHVGADAVMILPVSYWKLNEHEIFQHYRSVGDAITIPIMAYNNPA
MUL_1739|M.ulcerans_Agy99 RARYAQHVGADAVMILPVSYWKLNEHEIFQHYRSVGDAITIPIMAYNNPA
Mvan_0452|M.vanbaalenii_PYR-1 RGKYAQRAGADALMILPVSYWKLTDREIARHYASVAAATDLPIMAYNNPA
MSMEG_0877|M.smegmatis_MC2_155 NALAAQKAGADAVVATPPFYVAPHADEVIRHYRLIAAAVDVPVVAYDIPS
. **..:: . * : *: :. * :* : *: *
Mb2774c|M.bovis_AF2122/97 RSAVPIEPDTIRALASHPN-IVGVKDAKADLHSG-AQIMADTG--LAYYS
Rv2753c|M.tuberculosis_H37Rv RSAVPIEPDTIRALASHPN-IVGVKDAKADLHSG-AQIMADTG--LAYYS
MAV_3644|M.avium_104 RSVVPIESSTIRALASHPN-IVGVKDAKADLHAG-GQIIAETG--LAYYS
MLBr_01513|M.leprae_Br4923 RSVVPIEPDTIRALASHPN-IVGVKEAKADLYSG-ARIMADTG--LAYYS
Mflv_4000|M.gilvum_PYR-GCK RSVVPIEWDTIRRLAQHPN-IVAIKDAKADLHGG-GQIIAETG--LAYYS
TH_0834|M.thermoresistible__bu RSVVPLAWDTIRRLSEHRN-IVAIKDAKGDLHGA-AQIIAETG--LAYYS
MAB_3084c|M.abscessus_ATCC_199 RSVVPIDFHIIRELAAHPN-IVAVKDAKGDLHGG-AQIIAGTD--LVYYS
MMAR_2154|M.marinum_M TSGVDMRPELLVRMFESIDNLTMVKESTGDLSRM-RRIAELSGGRLPFYN
MUL_1739|M.ulcerans_Agy99 TSGVDMRPELLVRMFESIDNLTMVKESTGDLSRM-RRIAELSGGRLPFYN
Mvan_0452|M.vanbaalenii_PYR-1 TSGIDMSPELLVRMFTDIDNVAMVKESTGDLSRM-QRIKELTGGALPFYN
MSMEG_0877|M.smegmatis_MC2_155 ATHSPLTRSVVAQLAGEKT-VAALKDSSGDLDGFRAHLAANADTGLRVFT
: : : : :. :*::..** :: :. * :.
Mb2774c|M.bovis_AF2122/97 GDDALNLPWLAMGATGFISVIAHLAAGQLRELLSAFGSGDIATARKIN--
Rv2753c|M.tuberculosis_H37Rv GDDALNLPWLAMGATGFISVIAHLAAGQLRELLSAFGSGDIATARKIN--
MAV_3644|M.avium_104 GDDALNLPWLAMGATGFISVISHVAANQLRELLSAFTSGDIATARKIN--
MLBr_01513|M.leprae_Br4923 GDDALNLPWLAVGAIGFISVISHLAAGQLRELLSAFGSGDITTARKIN--
Mflv_4000|M.gilvum_PYR-GCK GDDALNLPWLAMGAVGFISVWGHLAASQLRDMLSAFASGDVATARKIN--
TH_0834|M.thermoresistible__bu GDDSLNLPWLAVGAVGFISVWGHLAAGQLRDMLTAFNSGDMTTARKIA--
MAB_3084c|M.abscessus_ATCC_199 GDDPLNLPWLSVGAVGFVSVISHFVAGPLRELLAAYNAGDVETARKLN--
MMAR_2154|M.marinum_M GSNPLVLDALKVGAAGWCTAAPNLWPQPCIDLYEAVCAGDLEKAQILY--
MUL_1739|M.ulcerans_Agy99 GSNPLVLDALKVGAAGWCTAAPNLWPQPCIDLYEAVCAGDLEKAQILY--
Mvan_0452|M.vanbaalenii_PYR-1 GSNPLVLDALTEGAAGWCTAAPNLRPQPCLDLYTAVRDGERSRAQEIY--
MSMEG_0877|M.smegmatis_MC2_155 GSETMTDVALNLGAHGAVPGLANVDPHGYVRIRQAARVGDWATAASEQER
*.:.: * ** * . :. . : * *: *
Mb2774c|M.bovis_AF2122/97 -------IAVAPLCNAMSRLGGVTLSKAGLRLQG-IDVGDPRLPQVAATP
Rv2753c|M.tuberculosis_H37Rv -------IAVAPLCNAMSRLGGVTLSKAGLRLQG-IDVGDPRLPQVAATP
MAV_3644|M.avium_104 -------ATVAPLCDAMARLGGVTMSKAGLRLQG-IDVGDPRLPQVPATP
MLBr_01513|M.leprae_Br4923 -------VAIGPLCSAMDRLGGVTMSKAGLRLQG-IDVGDPRLPQMPATA
Mflv_4000|M.gilvum_PYR-GCK -------VALGPLSAAQSRLGGVTLSKAGLRLQG-FEVGDPRLPQIPAND
TH_0834|M.thermoresistible__bu -------VTLSPLNAAQTRLGGVSMSKAGLRLLG-IEVGDPRLPQVPPGP
MAB_3084c|M.abscessus_ATCC_199 -------AQVAPITEACRRLGAVGAVKAGLRLQG-IEVGDPRLPNVPPSP
MMAR_2154|M.marinum_M -------DDLKPLLEFIVAGGLATTVKAGLGLLG-FPVGDPRPPLLPLDE
MUL_1739|M.ulcerans_Agy99 -------DDLKPLLEFIVAGRLATTVKAGLGLLG-FPVGDPRPPLLPLDE
Mvan_0452|M.vanbaalenii_PYR-1 -------DDLAPLLRFIVAGGLPTTVKAGLELLG-VSVGDPRAPLLALDD
MSMEG_0877|M.smegmatis_MC2_155 LRRIFAIVDVADRTRIGLTAGALGAFKAAQYLRGVIDTPRCADPLLPLES
: **. * * . . * :.
Mb2774c|M.bovis_AF2122/97 EQIDALAADMRAASVLR
Rv2753c|M.tuberculosis_H37Rv EQIDALAADMRAASVLR
MAV_3644|M.avium_104 EQIEALTADMRAASVLR
MLBr_01513|M.leprae_Br4923 EQIDELAVDMRAASVLR
Mflv_4000|M.gilvum_PYR-GCK VQLQALAADMRAASVLR
TH_0834|M.thermoresistible__bu EQLRLLADDMRAAAVLR
MAB_3084c|M.abscessus_ATCC_199 EQIEGLAADMRAAGVL-
MMAR_2154|M.marinum_M HGRAKLGRLLAGC----
MUL_1739|M.ulcerans_Agy99 HGRAKLGRLLAGC----
Mvan_0452|M.vanbaalenii_PYR-1 TGRAELKTLLAVA----
MSMEG_0877|M.smegmatis_MC2_155 SEIDAVRTILEREGVLG
: :