For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTTSTTLGGYVRDQLQTPLTLVGGFFRMCVLTGKALFRWPFQWREFILQCWFIMRVGFLPTIMVSIPLTV LLIFTLNILLAQFGAADISGSGAAIGAVTQLGPLTTVLVVAGAGSTAICADLGARTIREEIDAMEVLGID PIHRLVVPRVLASMLVATLLNGLVITVGLVGGFLFGVYLQNVSGGAYLATLTLITGLPEVVIATIKAATF GLIAGLVGCYRGLTVRGGSKGLGTAVNETVVLCVIALFAVNVILTTIGVRFGTGR
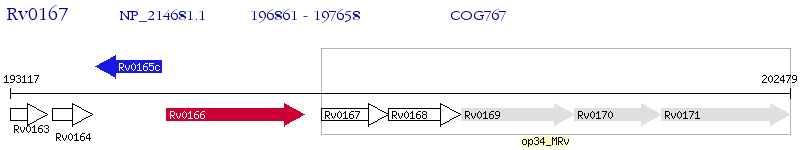
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0167 | yrbE1A | - | 100% (265) | CONSERVED HYPOTHETICAL INTEGRAL MEMBRANE PROTEIN YRBE1A |
| M. tuberculosis H37Rv | Rv0587 | yrbE2A | e-114 | 76.14% (264) | integral membrane protein YrbE2a |
| M. tuberculosis H37Rv | Rv3501c | yrbE4A | 3e-91 | 66.40% (247) | integral membrane protein YrbE4a |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0173 | yrbE1A | 1e-147 | 100.00% (265) | integral membrane protein YRBE1A |
| M. gilvum PYR-GCK | Mflv_0713 | - | 1e-122 | 81.44% (264) | hypothetical protein Mflv_0713 |
| M. leprae Br4923 | MLBr_02587 | yrbE1A | 1e-130 | 88.35% (266) | hypothetical protein MLBr_02587 |
| M. abscessus ATCC 19977 | MAB_4155c | - | 2e-89 | 64.40% (250) | hypothetical protein MAB_4155c |
| M. marinum M | MMAR_0410 | yrbE1A | 1e-132 | 88.72% (266) | integral membrane protein YrbE1A |
| M. avium 104 | MAV_5017 | - | 1e-136 | 92.97% (256) | hypothetical protein MAV_5017 |
| M. smegmatis MC2 155 | MSMEG_0132 | - | 1e-117 | 77.44% (266) | hypothetical protein MSMEG_0132 |
| M. thermoresistible (build 8) | TH_0155 | - | 1e-122 | 82.64% (265) | CONSERVED HYPOTHETICAL INTEGRAL MEMBRANE PROTEIN YRBE4A |
| M. ulcerans Agy99 | MUL_1060 | yrbE1A | 1e-131 | 88.35% (266) | integral membrane protein YrbE1A |
| M. vanbaalenii PYR-1 | Mvan_0132 | - | 1e-123 | 83.71% (264) | hypothetical protein Mvan_0132 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0713|M.gilvum_PYR-GCK --MTTTSGLSGYVREQVKPGLLAVGGFVRMCVLVGKATFRTPFQWREFIL
Mvan_0132|M.vanbaalenii_PYR-1 --MTSSSNLTGYVREQVKPGLEAVGGFVRMCVLVGKATLRPPFQWREFIL
TH_0155|M.thermoresistible__bu --VTTTDGLVGYVRGQAKPVLESVGGFVRMTVLTGKALFKPPFQWREFIL
MSMEG_0132|M.smegmatis_MC2_155 -MTASTDGFVDYLRGQLEKPLATVGGFFKMSVMTGKALFTRPFQWKEFVL
Rv0167|M.tuberculosis_H37Rv MTTST--TLGGYVRDQLQTPLTLVGGFFRMCVLTGKALFRWPFQWREFIL
Mb0173|M.bovis_AF2122/97 MTTST--TLGGYVRDQLQTPLTLVGGFFRMCVLTGKALFRWPFQWREFIL
MMAR_0410|M.marinum_M MTTSSGPHLMGYLRDQLETPLTMVGGFFRMCVLTGKALFRRPFQWRELIL
MUL_1060|M.ulcerans_Agy99 MTTSSGPHLMGYLRDQLETSLTMVGGFFRMCVLTGKALFRRPFQWRELIL
MAV_5017|M.avium_104 ---------MGYLRDQLETPLTLVGGFFRMCVLTGKALFRWPFQWREFVL
MLBr_02587|M.leprae_Br4923 MTTETRSSLVEYAHTKLQTPLVLVGGFFRMLVLTGKALFRRPFQWREFIL
MAB_4155c|M.abscessus_ATCC_199 ------------MKQQLAAPARAVGGFVSMSLATFRNIFRRPFQGQEFLD
: : ****. * : . : : *** :*::
Mflv_0713|M.gilvum_PYR-GCK QSWFLLRVAFLPTVAVSIPLTVLLIFTLNILLTEFGAADISGAGAALGAV
Mvan_0132|M.vanbaalenii_PYR-1 QSWFLMRVAFLPTLAVSIPLTVLLIFTLNILLAEFGAADVSGAGAAIGAV
TH_0155|M.thermoresistible__bu QAWFLFRVSFLPTIAVSIPLTVLIIFTLNILLAEFGAADVSGAGAALGAV
MSMEG_0132|M.smegmatis_MC2_155 QSWFLIRVAFLPTLAVSIPLTVLIIFTLNILLAEFGAADVSGAGAALGAV
Rv0167|M.tuberculosis_H37Rv QCWFIMRVGFLPTIMVSIPLTVLLIFTLNILLAQFGAADISGSGAAIGAV
Mb0173|M.bovis_AF2122/97 QCWFIMRVGFLPTIMVSIPLTVLLIFTLNILLAQFGAADISGSGAAIGAV
MMAR_0410|M.marinum_M QCWFVMRVALLPTIMVSIPLTVLLIFTLNVLLAQFGAADISGAGAAIGAV
MUL_1060|M.ulcerans_Agy99 QCWFVMRVALLPTIMVSIPLTVLLIFTLNVLLAQFGAADISGAGAAIGAV
MAV_5017|M.avium_104 QCWFIMRVAFLPTIMVSIPLTVLLIFTLNVLLAQFGAADLSGAGAAIGAV
MLBr_02587|M.leprae_Br4923 QCWFIMRVAFLPTVMVSIPLTVLLIFTLNVLLAQFSAADLSGAGAAIGAV
MAB_4155c|M.abscessus_ATCC_199 QTWFIARVSLLPTLLVAIPFTVLVAFTLNILLREIGAADLSGAATAFGTV
* **: **.:***: *:**:***: ****:** ::.***:**:.:*:*:*
Mflv_0713|M.gilvum_PYR-GCK TQLGPLVTVLVVAGAGSTAICADLGARTIREEIDALEVLGIDPIHRLVVP
Mvan_0132|M.vanbaalenii_PYR-1 TQLGPLVTVLVVAGAGSTAICADLGARTIREEIDALEVLGIDPIHRLVVP
TH_0155|M.thermoresistible__bu TQLGPLVTVLVVAGAGSTAICADLGARTIREEIDALEVLGIDPIHRLVVP
MSMEG_0132|M.smegmatis_MC2_155 TQLGPLVTVLVVAGAGSTAICADLGARTVREEIDALEVLGIDPIERLVVP
Rv0167|M.tuberculosis_H37Rv TQLGPLTTVLVVAGAGSTAICADLGARTIREEIDAMEVLGIDPIHRLVVP
Mb0173|M.bovis_AF2122/97 TQLGPLTTVLVVAGAGSTAICADLGARTIREEIDAMEVLGIDPIHRLVVP
MMAR_0410|M.marinum_M TQLGPLTTVLVVAGAGSTAICADLGARTIREEIDAMEVLGIDPIHRLVVP
MUL_1060|M.ulcerans_Agy99 TQLGPLTTVLVVAGAGSTAICADLGARTIREEIDAMEVLGIDPIHRLVVP
MAV_5017|M.avium_104 TQLGPLTTVLVVAGAGSTAICADLGARTIREEIDAMEVLGIDPIHRLVVP
MLBr_02587|M.leprae_Br4923 TQLGPLTTVLVVAGAGSTSICADLGARTIREEIDAMEVLGIDPIHRLVVP
MAB_4155c|M.abscessus_ATCC_199 TQLGPVVTVLVVAGAGATAICADLGARTIREEIDAMQVLGIDPIQRLVVP
*****:.*********:*:*********:******::*******.*****
Mflv_0713|M.gilvum_PYR-GCK RVIASTFVAMLLNGAVITIGLVGGFIFGVYLQNVSAGAYVSTLTLVTGLP
Mvan_0132|M.vanbaalenii_PYR-1 RVVASTFVAILLNGAVITVGLVGGFIFGVYLQNVSAGAYVSTLTLITGLP
TH_0155|M.thermoresistible__bu RVVASTFVALLLNGAVITIGLVGGFVFGVYVQNVSAGAYLSTLTLVTGLP
MSMEG_0132|M.smegmatis_MC2_155 RVVASTFVAFMLNGAVITIGLVGGFFFGVYIQNVSAGAYVSTLTLLTGFP
Rv0167|M.tuberculosis_H37Rv RVLASMLVATLLNGLVITVGLVGGFLFGVYLQNVSGGAYLATLTLITGLP
Mb0173|M.bovis_AF2122/97 RVLASMLVATLLNGLVITVGLVGGFLFGVYLQNVSGGAYLATLTLITGLP
MMAR_0410|M.marinum_M RVIAATFVATLLNALVITVGLVGGYLFGVYLQNINGGAYLATLTTITGMP
MUL_1060|M.ulcerans_Agy99 RVIAATFVATLLNALVITVGLVGGYLFGVYLQNINGGAYLATLTTITGMP
MAV_5017|M.avium_104 RVIAATLVATLLNGLVITVGLVGGYLFGVYLQNVSGGAYLATLTTITGLP
MLBr_02587|M.leprae_Br4923 RVLAATLVATLLNGLVITVGLVGGYLFGVYLQNVSGGAYLATLTTITGLP
MAB_4155c|M.abscessus_ATCC_199 RVLASTFVALLLNGLVCAIGMVGGYVFSVFLQGVNPGAFINGLTVLTGLG
**:*: :** :**. * ::*:***:.*.*::*.:. **:: ** :**:
Mflv_0713|M.gilvum_PYR-GCK EVLISLVKALTFGLIAGLVGCYRGLTVAGGAKGVGTAVNETLVLCVIALF
Mvan_0132|M.vanbaalenii_PYR-1 EVVISIVKAATFGLIAGLVGCYRGLTVAGGAKGVGTAVNETLVLCVIALF
TH_0155|M.thermoresistible__bu EVVISIVKATTFGLIAGLVGCYRGLTVSGGAKGVGTAVNETLVLCVIALF
MSMEG_0132|M.smegmatis_MC2_155 EVLISVVKATLFGMIAGLVGCYRGLTVAGGSKGVGTAVNETLVLCVVALF
Rv0167|M.tuberculosis_H37Rv EVVIATIKAATFGLIAGLVGCYRGLTVRGGSKGLGTAVNETVVLCVIALF
Mb0173|M.bovis_AF2122/97 EVVIATIKAATFGLIAGLVGCYRGLTVRGGSKGLGTAVNETVVLCVIALF
MMAR_0410|M.marinum_M EVTIAMVKAATFGLIAGLVGCYRGLTVRGGSKGLGTAVNETVVLCVVALY
MUL_1060|M.ulcerans_Agy99 EVTIAMVKAATFGLIAGLVGCYRGLTVRGGSKGLGTAVNETVVLCVVALY
MAV_5017|M.avium_104 EVVIATVKAAVFGLIAGLVGCFRGLTVRGGSKGLGTAVNETVVLCVVALY
MLBr_02587|M.leprae_Br4923 EVVIATVKAATFGLIAGLVGCYRGLIVRGGSNGLGAAVNETVVLCVVALY
MAB_4155c|M.abscessus_ATCC_199 ELMISEVKAFLFGIFAGLVGCYRGLTVKGGPKGVGDAVNETVVYAFICLF
*: *: :** **::******:*** * **.:*:* *****:* ..:.*:
Mflv_0713|M.gilvum_PYR-GCK AVNVVLTTIGVRFGTGT
Mvan_0132|M.vanbaalenii_PYR-1 AVNVVLTTIGVRFGTGS
TH_0155|M.thermoresistible__bu AVNVILTTIGVRFGTGR
MSMEG_0132|M.smegmatis_MC2_155 AVNVVLTTIGVRFGTGR
Rv0167|M.tuberculosis_H37Rv AVNVILTTIGVRFGTGR
Mb0173|M.bovis_AF2122/97 AVNVILTTIGVRFGTGR
MMAR_0410|M.marinum_M AVNVILTTIGVRFGTGH
MUL_1060|M.ulcerans_Agy99 AVNVILTTIGVRFGTGH
MAV_5017|M.avium_104 AVNVVLTTIGVRFGTGH
MLBr_02587|M.leprae_Br4923 AVNVVLTTIGVRFGTGH
MAB_4155c|M.abscessus_ATCC_199 VINVVMTAIGVRVLVKH
.:**::*:****. .