For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PAKVAAMRMTRVLGAVLAAVVLTTAGCARVVEGTPQANTDPPGSEITEDGSGILIGYHAAPVRIEIFAEP QCPPCGRLQRDYGDEIAEYVGEGRLAVVYRPMTFLDLDGPGYSAHVSNALFLAAGPDTDATTFQKFVQTV WSNQEPEGSAGPSDDELADMASESGISADLVDRIRSGDEGVDIDMMAEQNIQYLTDAAGVAATPAVYDLN EDTLVPLRDDWLSDLMLGLGS*
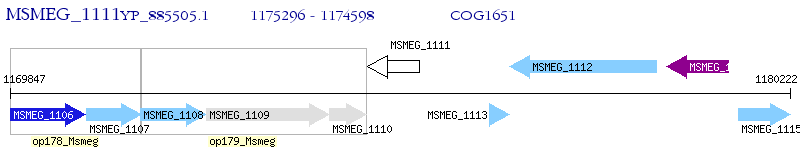
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1111 | - | - | 100% (232) | hypothetical protein MSMEG_1111 |
| M. smegmatis MC2 155 | MSMEG_2410 | - | 8e-12 | 24.51% (204) | putative serine-threonine protein kinase |
| M. smegmatis MC2 155 | MSMEG_1215 | - | 1e-11 | 29.82% (171) | serine/threonine-protein kinase PknE, putative |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1772 | pknE | 9e-14 | 28.23% (209) | transmembrane serine/threonine-protein kinase E |
| M. gilvum PYR-GCK | Mflv_5261 | - | 8e-47 | 43.36% (226) | hypothetical protein Mflv_5261 |
| M. tuberculosis H37Rv | Rv1743 | pknE | 9e-14 | 28.23% (209) | transmembrane serine/threonine-protein kinase E |
| M. leprae Br4923 | MLBr_01667 | - | 2e-07 | 23.04% (217) | hypothetical protein MLBr_01667 |
| M. abscessus ATCC 19977 | MAB_3089c | - | 7e-23 | 32.08% (212) | hypothetical protein MAB_3089c |
| M. marinum M | MMAR_2581 | pknE | 5e-14 | 28.57% (231) | transmembrane serine/threonine-protein kinase E PknE |
| M. avium 104 | MAV_3813 | - | 1e-07 | 25.49% (153) | hypothetical protein MAV_3813 |
| M. thermoresistible (build 8) | TH_2711 | - | 2e-36 | 41.26% (223) | PUTATIVE putative conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_3180 | pknE | 5e-14 | 30.56% (180) | transmembrane serine/threonine-protein kinase E PknE |
| M. vanbaalenii PYR-1 | Mvan_0977 | - | 1e-40 | 41.85% (227) | hypothetical protein Mvan_0977 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1772|M.bovis_AF2122/97 -MDGTAESREGTQFGPYRLRRLVGRGGMGDVYEAEDTVRERIVALKLMSE
Rv1743|M.tuberculosis_H37Rv -MDGTAESREGTQFGPYRLRRLVGRGGMGDVYEAEDTVRERIVALKLMSE
MMAR_2581|M.marinum_M MSDATADSREGSQFGPYRLRRIVGRGAMGDVYEAEDTVRERVVALKLMSQ
MUL_3180|M.ulcerans_Agy99 MSDATADSREGSQFGPYRLRRIVGRGAMGDVYEAEDTVRERVVALKLMSQ
Mflv_5261|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0977|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2711|M.thermoresistible__bu --------------------------------------------------
MSMEG_1111|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01667|M.leprae_Br4923 --------------------------------------------------
MAV_3813|M.avium_104 --------------------------------------------------
MAB_3089c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1772|M.bovis_AF2122/97 TLSSDPDFRTRMQREARTAGRLQEPHVVPIHDFGEIDGQLYVDMRLINGV
Rv1743|M.tuberculosis_H37Rv TLSSDPVFRTRMQREARTAGRLQEPHVVPIHDFGEIDGQLYVDMRLINGV
MMAR_2581|M.marinum_M TLSNDPVFRTRMQREARTAGRLQEPHVVPIHDYGEIDGQLYVDMRLIDGK
MUL_3180|M.ulcerans_Agy99 TLSNDPVFRTRMQREARTAGRLQEPHVVPIHDYGEIDGQLYVDMRLIDGK
Mflv_5261|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0977|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2711|M.thermoresistible__bu --------------------------------------------------
MSMEG_1111|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01667|M.leprae_Br4923 --------------------------------------------------
MAV_3813|M.avium_104 --------------------------------------------------
MAB_3089c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1772|M.bovis_AF2122/97 DLAAMLRRQGPLAPPRAVAIVRQIGSALDAAHAAGATHRDVKPENILVSA
Rv1743|M.tuberculosis_H37Rv DLAAMLRRQGPLAPPRAVAIVRQIGSALDAAHAAGATHRDVKPENILVSA
MMAR_2581|M.marinum_M DLAAMLKRYGPLPPPRAVAITRQIGSALDAAHAAGVTHRDVKPENILVSH
MUL_3180|M.ulcerans_Agy99 DLAAMLKRYGPLPPPRAVAITRQIGSALDAAHAAGVTHRDVKPENILVSH
Mflv_5261|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0977|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2711|M.thermoresistible__bu --------------------------------------------------
MSMEG_1111|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01667|M.leprae_Br4923 --------------------------------------------------
MAV_3813|M.avium_104 --------------------------------------------------
MAB_3089c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1772|M.bovis_AF2122/97 DDFAYLVDFGIASATTDEKLTQLGNTVGTLYYMAPERFSESHATYRADIY
Rv1743|M.tuberculosis_H37Rv DDFAYLVDFGIASATTDEKLTQLGNTVGTLYYMAPERFSESHATYRADIY
MMAR_2581|M.marinum_M DDFAYLVDFGIASATSDEKLTQLGSTVGTLYYMAPERFSSGDVTYRADIY
MUL_3180|M.ulcerans_Agy99 DDFAYLVDFGIASATSDEKLTQLGSTVGTLYYMAPERFSSGDVTYRADIY
Mflv_5261|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0977|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2711|M.thermoresistible__bu --------------------------------------------------
MSMEG_1111|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01667|M.leprae_Br4923 --------------------------------------------------
MAV_3813|M.avium_104 --------------------------------------------------
MAB_3089c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1772|M.bovis_AF2122/97 ALTCVLYECLTGSPPYQGDQLSVMGAHINQAIPRPSTVRPGIPVAFDAVI
Rv1743|M.tuberculosis_H37Rv ALTCVLYECLTGSPPYQGDQLSVMGAHINQAIPRPSTVRPGIPVAFDAVI
MMAR_2581|M.marinum_M ALACVLYECLTGSPPYRGDQLSVMGAHLSQPIPRPSAARPGIPAGFDNVI
MUL_3180|M.ulcerans_Agy99 ALACVLYECLTGSPPYRGDQLSVIGAHLSQPIPRPSAARPGIPAGFDNVI
Mflv_5261|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0977|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2711|M.thermoresistible__bu --------------------------------------------------
MSMEG_1111|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01667|M.leprae_Br4923 --------------------------------------------------
MAV_3813|M.avium_104 --------------------------------------------------
MAB_3089c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1772|M.bovis_AF2122/97 ARGMAKNPEDRYVTCGDLSAAAHAALATADQDRATDILRRSQVAKLPV--
Rv1743|M.tuberculosis_H37Rv ARGMAKNPEDRYVTCGDLSAAAHAALATADQDRATDILRRSQVAKLPV--
MMAR_2581|M.marinum_M ARGMAKDPADRYATCGDLSAAAYSALATPDQDRATDILQRSQVAQLPAGF
MUL_3180|M.ulcerans_Agy99 ARGMAKDPADRYATCGDLSAAAYSALATPDQDRATDILQRSQVAQLPAGF
Mflv_5261|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0977|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2711|M.thermoresistible__bu --------------------------------------------------
MSMEG_1111|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01667|M.leprae_Br4923 --------------------------------------------------
MAV_3813|M.avium_104 --------------------------------------------------
MAB_3089c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1772|M.bovis_AF2122/97 PSTHP--------------VSPGTRWPQP--------------TPWAG--
Rv1743|M.tuberculosis_H37Rv PSTHP--------------VSPGTRWPQP--------------TPWAG--
MMAR_2581|M.marinum_M ASTPPGGFAPSSPAVPVPGSAPSTGWPAPSVPGAPVAPSAPVPTPWAGVP
MUL_3180|M.ulcerans_Agy99 ASTSPGGFAPSSPAVPVPGSAPSTGWPAPSVPGAPVAPSAPVPTPWAGVP
Mflv_5261|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0977|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2711|M.thermoresistible__bu --------------------------------------------------
MSMEG_1111|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01667|M.leprae_Br4923 --------------------------------------------------
MAV_3813|M.avium_104 --------------------------------------------------
MAB_3089c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1772|M.bovis_AF2122/97 ----------GAPPWGPPSSPLPRSARQPWLWVGVAVAVVVALAGGLGIA
Rv1743|M.tuberculosis_H37Rv ----------GAPPWGPPSSPLPRSARQPWLWVGVAVAVVVALAGGLGIA
MMAR_2581|M.marinum_M GWGVGHTGTTGAPPWGQPP-PQPTVAAKTWPWIAAAVAVVVVIAGVLGVV
MUL_3180|M.ulcerans_Agy99 GWGVGHTGTTGAPPWGQPP-PEPTVAAKTWPWIAAAVAVVVVIAGVLGVV
Mflv_5261|M.gilvum_PYR-GCK ----------------------------------MRISRPLALVAALG-L
Mvan_0977|M.vanbaalenii_PYR-1 ----------------------------------MHWIRPLAALSAFGLL
TH_2711|M.thermoresistible__bu -------------------------------------MRWLTLTAMLGVA
MSMEG_1111|M.smegmatis_MC2_155 ---------------------------MPAKVAAMRMTRVLG-AVLAAVV
MLBr_01667|M.leprae_Br4923 -----MRISEPVADNRKRLQRFDLKSVDG-RSGRAVVIAGTVIVAIFAVV
MAV_3813|M.avium_104 -----------MADNPKRPPRFDMKSASGGKSGRLVQIGGTAFVVIFAVA
MAB_3089c|M.abscessus_ATCC_199 ------------------------------MSAPGAAGVPKVLLLAAIVL
Mb1772|M.bovis_AF2122/97 LAHPWRSSGPRTSAPPPPPPADAVELRVLNDG--VFVGSSVAPTTIDIFN
Rv1743|M.tuberculosis_H37Rv LAHPWRSSGPRTSAPPPPPPADAVELRVLNDG--VFVGSSVAPTTIDIFN
MMAR_2581|M.marinum_M IAHPWRSPGPPTASAPPPPPPDAVQLRVLNDG--VFIGSSAATTTIDIFN
MUL_3180|M.ulcerans_Agy99 IAHPWRSPGPPTASAPPPPPPDAVQLRVLNDG--VFIGSSAATTTIDIFN
Mflv_5261|M.gilvum_PYR-GCK LVGPGCSRAVTGTPRADPAP---VPLAVAEDGFGVVAGFDTAPAKIEIYT
Mvan_0977|M.vanbaalenii_PYR-1 LTAVGCAKQVTGVARVDATG---APLAVTEDGFGIVAGFDDAPVKIEIYT
TH_2711|M.thermoresistible__bu VLLSGCTVTVDGAASAPADAPGDATVVVAEDGYGVELGYQFAPARIEIYI
MSMEG_1111|M.smegmatis_MC2_155 LTTAGCARVVEGTPQANTDP---PGSEITEDGSGILIGYHAAPVRIEIFA
MLBr_01667|M.leprae_Br4923 VVFAVMKSQHAKQDGAAPLTGSGDTVRVTSSKLVTQPGTHDPKVVMSFYE
MAV_3813|M.avium_104 LVFYIVTSHHKK----AGPTGAGDTVRVTSSKLITQPGSSNPKAVVTLYE
MAB_3089c|M.abscessus_ATCC_199 LVLTGCARTVEGTASAPVEA---AGWGNSQGAGVTVGGAGDVAAGITVFL
: .. * . : .:
Mb1772|M.bovis_AF2122/97 EPICPPCGSFIRSYASDIDTAVADKQLAVRYHLLNFLDDQSHSKNYSTRA
Rv1743|M.tuberculosis_H37Rv EPICPPCGSFIRSYASDIDTAVADKQLAVRYHLLNFLDDQSHSKNYSTRA
MMAR_2581|M.marinum_M EPICPPCGAFIRSYASDIDAAVANKKLAVRYHLLNFLDEQSHTKTYSTRA
MUL_3180|M.ulcerans_Agy99 EPICPPCGAFIRSYASDIDAAVANKKLAVRYHLLNFLDEQSHTKTYSTRA
Mflv_5261|M.gilvum_PYR-GCK EPQCTHCGDLQREYGDELAYHITVGDLQVVYRPLTFLDDSY--DGYSATV
Mvan_0977|M.vanbaalenii_PYR-1 EPQCTHCSDLQHEFGEQLADSISVGTLQVTYRPLTFLDDDY--DGYSAKV
TH_2711|M.thermoresistible__bu EPLCPACARLQRRYGEDLRSAIERDELRVTYRPLTFLDRS---GGYSKRV
MSMEG_1111|M.smegmatis_MC2_155 EPQCPPCGRLQRDYGDEIAEYVGEGRLAVVYRPMTFLDLDG--PGYSAHV
MLBr_01667|M.leprae_Br4923 DFLCPGCGDFERNFGPTVSKLIDIGAIAADYSVVSILDHPRN-HNYPSRA
MAV_3813|M.avium_104 DFLCPACGNFERTFGPTVSKLIDLGAIAADYSMVSILDSPRN-QNYSSRA
MAB_3089c|M.abscessus_ATCC_199 DFQCPFCQRFEAQYGEDITTYVTEGRLQVTYRPVSFLDRISASGDYSSRA
: *. * : :. : : : . * :.:** *. .
Mb1772|M.bovis_AF2122/97 VAASYCVAGQNDPKLYASFYSALFGSDFQPQENAASDR--TDAELAHLAQ
Rv1743|M.tuberculosis_H37Rv VAASYCVAGQNDPKLYASFYSALFGSDFQPQENAASDR--TDAELAHLAQ
MMAR_2581|M.marinum_M VAASYCVAAQDDPKVYTDFYAALFASDFQPQEAAASDR--TDAELAHLAQ
MUL_3180|M.ulcerans_Agy99 VAASYCVAAQDDPKVYTDFYAALFASDFQPQEAAASDR--TDAELAHLAQ
Mflv_5261|M.gilvum_PYR-GCK ANALFAATEAKGEFPANGTQFQRFVEQLWVNQDPGGQP-FAADELRRMAD
Mvan_0977|M.vanbaalenii_PYR-1 ANALFLATEAVDNSAATGTQLQRFVEELWINQDPGGAV-FTADELRDMAV
TH_2711|M.thermoresistible__bu SNALFAAAEPGSD--AGATQVQAFVMQTYRLLSPVGDN-LDARGLAAVAG
MSMEG_1111|M.smegmatis_MC2_155 SNALFLAAGPDTD----ATTFQKFVQTVWSNQEPEGSAGPSDDELADMAS
MLBr_01667|M.leprae_Br4923 GAASLCVADESMD-AYRRFRAALYSRSFQPSELASSFP--ENAKLIEIAR
MAV_3813|M.avium_104 GAAALCVADESLD-AFRRFHTALFSTAIQPSETGKTFP--DNARLIELAR
MAB_3089c|M.abscessus_ATCC_199 AAALFLIDKAGAT----DAVILGFIGEMFRRQPVEGVGNLTNLQISEIAA
* : : :*
Mb1772|M.bovis_AF2122/97 TVGAEPTAISCIKSG---ADLGTAQTKATNASETLAGFNASGT----PFV
Rv1743|M.tuberculosis_H37Rv TVGAEPTAISCIKSG---ADLGTAQTKATNASETLAGFNASGT----PFV
MMAR_2581|M.marinum_M TVGANGTATSCIKAG---NDMGTARTKAAAADATLSELNASGT----PFV
MUL_3180|M.ulcerans_Agy99 TVGANGTATSCIKAG---NDMGTARTKAAAADATLSELNAG---------
Mflv_5261|M.gilvum_PYR-GCK DAGLPGPVADRVAGG---SEAVDLVDMEDHNFEMLFEVDQITTGTPTVYD
Mvan_0977|M.vanbaalenii_PYR-1 SAGLPEAVADHVATD---SEAVDVAEMDDTNFGLLFDIDRVDTGTPTVFD
TH_2711|M.thermoresistible__bu DVGLPDDVVDLIADN---APAVDVEAMDSYNTERFEELTGKRAYTPMVFD
MSMEG_1111|M.smegmatis_MC2_155 ESGISADLVDRIRSG---DEGVDIDMMAEQNIQYLTDAAGVAA-TPAVYD
MLBr_01667|M.leprae_Br4923 EAGVVGKVPDCINSGKYLAKVTAEAALAKISATPTIKINGDEYDPLTPEA
MAV_3813|M.avium_104 ESGVVGKVPDCINSGKYIAKVTGEAAAAKIRATPTIKINGEDYDPSTPDA
MAB_3089c|M.abscessus_ATCC_199 GAGVTGEVLREISSL----QVNEVDRQRTAGNEQQLTDHGLGG--VPGVI
* :
Mb1772|M.bovis_AF2122/97 WDGSMVVNYQDPSWLARLIG----
Rv1743|M.tuberculosis_H37Rv WDGSMVVNYQDPSWLARLIG----
MMAR_2581|M.marinum_M WDGSKSIDLQNPSWLTKLIG----
MUL_3180|M.ulcerans_Agy99 --------------LRRC------
Mflv_5261|M.gilvum_PYR-GCK LNAAEKLDIADSSWLADLVKS---
Mvan_0977|M.vanbaalenii_PYR-1 LDAGEKLDIFDDEWLSSLLRSH--
TH_2711|M.thermoresistible__bu LNARAEVDVDDPDWVADVIEGS--
MSMEG_1111|M.smegmatis_MC2_155 LNEDTLVPLRD-DWLSDLMLGLGS
MLBr_01667|M.leprae_Br4923 LVAKIKAIVGAVPGIDVAVTATTT
MAV_3813|M.avium_104 LVGKIKEIVGDIPGLDGAVAPAAM
MAB_3089c|M.abscessus_ATCC_199 DNSGEMIDPSDSGWLSRLVGAR--
: