For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTDSALERTTDTVAQTIPQVRAQVDCGEITPVGLVRRALARISHVEDKIQAFAAVFADEALAVAQESAPQ AGPLWGIPVAVKDIYDVAGYRTGNGSLGAPEHLALRDAEAVRRLRDAGAIILGKATTHEYAYGVTTPPTR NPWSLDRIPGGSSGGTGAAIAAGVVAAGLGTDTGGSIRIPAALCGVTGHKPTFGLVSRSGVSALSSTLDH TGPLGRTVDDTVALLEVIAGHDPADPYSSSAPVPDFRAEFDRGLAGLTIGVAEPYFCDRLTPDVAAAFEE AIHTLERGGATITSVTFTDVDLCPRVVDVVCGVEAAAWHAAQIGMAPEKFSPEVRDALRLGASYTGVEYL EARRARCAVIAGMNALFDSGPDLLISPTIAMTAPPYGSTHVDLGGRTVGVLDGVNALTVPANVTGMPALT VPAGFGSDGLPIGLQLMGRPGADATVLGAGRAFERIAGFVDFTPNL
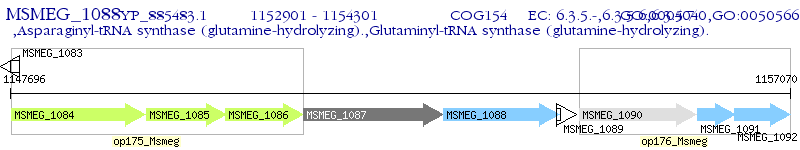
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1088 | gatA | - | 100% (466) | glutamyl-tRNA(Gln)/aspartyl-tRNA(Asn) amidotransferase, A subunit |
| M. smegmatis MC2 155 | MSMEG_1090 | - | 4e-95 | 41.83% (447) | amidase |
| M. smegmatis MC2 155 | MSMEG_4062 | - | 3e-64 | 41.88% (394) | glutamyl-tRNA(Gln) amidotransferase subunit A |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3036c | gatA | 1e-60 | 34.89% (493) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. gilvum PYR-GCK | Mflv_4246 | gatA | 9e-58 | 34.38% (480) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. tuberculosis H37Rv | Rv3011c | gatA | 1e-60 | 34.89% (493) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. leprae Br4923 | MLBr_01702 | gatA | 2e-56 | 33.96% (480) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. abscessus ATCC 19977 | MAB_3341c | gatA | 3e-61 | 34.01% (491) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. marinum M | MMAR_1702 | gatA | 1e-60 | 34.35% (492) | glutamyl-tRNA (Gln) amidotransferase (subunit A), GatA |
| M. avium 104 | MAV_3859 | gatA | 4e-59 | 34.17% (480) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. thermoresistible (build 8) | TH_2639 | - | 1e-61 | 35.09% (493) | PUTATIVE glutamyl-tRNA (Gln) amidotransferase (subunit A), GatA |
| M. ulcerans Agy99 | MUL_1939 | gatA | 1e-60 | 34.35% (492) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. vanbaalenii PYR-1 | Mvan_2115 | gatA | 2e-54 | 32.80% (497) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_01702|M.leprae_Br4923 ----MIGSLHEIIRLDASTLAAKIVAKELSSVEITQACLDQIEATDDTYR
MAV_3859|M.avium_104 --------MNEIIRSDAATLAARIAAKELSSVEATQACLDQIAATDERYH
Mb3036c|M.bovis_AF2122/97 --------MTDIIRSDAATLAAKIAIKEVSSTEITRACLDQIEATDETYH
Rv3011c|M.tuberculosis_H37Rv --------MTDIIRSDAATLAAKIAIKEVSSAEITRACLDQIEATDETYH
MMAR_1702|M.marinum_M --------MTDLIRSDAATLAAKIAAKEVSATELTQACLDQIEATDDRYH
MUL_1939|M.ulcerans_Agy99 --------MTDLIRSDAATLAAKIAAKEVSATELTQACLDQIEATDDRYH
Mflv_4246|M.gilvum_PYR-GCK --------MSELIKLDAATLADKISSKEVSSAEVTQAFLDQIAATDGDYH
Mvan_2115|M.vanbaalenii_PYR-1 --------MTDLIKLDAATLAAKIAAREVSATEVTQACLDQIAATDAEYH
MAB_3341c|M.abscessus_ATCC_199 --------MTDLIRQDAAALAALIHSGEVSSVEVTRAHLDQIASTDETYR
TH_2639|M.thermoresistible__bu --MSQGKSTDDLIRLDAATLGAKIAAKEISSTELTQACLDRIAATDDRYN
MSMEG_1088|M.smegmatis_MC2_155 MTDSALERTTDTVAQTIPQVRAQVDCGEITPVGLVRRALARISHVEDKIQ
: : . : : *::.. .: * :* .: .
MLBr_01702|M.leprae_Br4923 AFLHVAAEKALSAAAAVDK----AVAAGGQLSSTLAGVPLALKDVFTTVD
MAV_3859|M.avium_104 AFLHVAADEALGAAAVVDE----AVAAGERLPSPLAGVPLALKDVFTTVD
Mb3036c|M.bovis_AF2122/97 AFLHVAADEALAAAAAVDK----QVAAGEPLPSALAGVPLALKDVFTTSD
Rv3011c|M.tuberculosis_H37Rv AFLHVAADEALAAAAAIDK----QVAAGEPLPSALAGVPLALKDVFTTSD
MMAR_1702|M.marinum_M AFLHIGAHEALSAAAAVDT----ALAAGERLPSALAGVPLALKDVFTTVD
MUL_1939|M.ulcerans_Agy99 AFLHIGAHEALSAAAAVDT----ALAAGERLPSALAGVPLALKDVFTTVD
Mflv_4246|M.gilvum_PYR-GCK AFLHVGAEQALAAAQTVDR----AVAAGEHLPSPLAGVPLALKDVFTTTD
Mvan_2115|M.vanbaalenii_PYR-1 AFLHVAGDQALAAAATVDKAIHEATAAGERLPSPLAGVPLALKDVFTTTD
MAB_3341c|M.abscessus_ATCC_199 AFLHVAGEQALAAAKDVDD----AIAAGS-VPSGLAGVPLALKDVFTTRD
TH_2639|M.thermoresistible__bu AFLHVAANKALSAAARVDA----AVAAGEELPSPLAGVPLALKDVFTTID
MSMEG_1088|M.smegmatis_MC2_155 AFAAVFADEALAVAQESAP-----------QAGPLWGIPVAVKDIYDVAG
** : ..:**..* .. * *:*:*:**:: . .
MLBr_01702|M.leprae_Br4923 MPTTCGSKILQGWHSPYDATVTTRLRAAGIPILGKTNMDEFAMGSSTENS
MAV_3859|M.avium_104 MPTTCGSKILQGWRSPYDATVTTKLRAAGIPILGKTNMDEFAMGSSTENS
Mb3036c|M.bovis_AF2122/97 MPTTCGSKILEGWRSPYDATLTARLRAAGIPILGKTNMDEFAMGSSTENS
Rv3011c|M.tuberculosis_H37Rv MPTTCGSKILEGWRSPYDATLTARLRAAGIPILGKTNMDEFAMGSSTENS
MMAR_1702|M.marinum_M MPTTCGSKILEGWRSPYDATLTLRLRAAGIPILGKTNMDEFAMGSSTENS
MUL_1939|M.ulcerans_Agy99 MPTTCGSKILEGWRSPYDATLTLRLRAAGIPILGKTNMDEFAMGSSTENS
Mflv_4246|M.gilvum_PYR-GCK MPTTCGSKVLEGWTSPYDATVTAKLRSAGIPILGKTNMDEFAMGSSTENS
Mvan_2115|M.vanbaalenii_PYR-1 MPTTCGSKILEGWTSPYDATVTAKLRAAGIPILGKTNMDEFAMGSSTENS
MAB_3341c|M.abscessus_ATCC_199 MPTTCGSKILENWVPPYDATVTARLRGAGIPILGKTNMDEFAMGSSTENS
TH_2639|M.thermoresistible__bu MPTTCGSKILDGWQAPYDATVTVALRAAGIPILGKTNLDEFAMGSSTENS
MSMEG_1088|M.smegmatis_MC2_155 YRTGNGSLGAPEHLALRDAEAVRRLRDAGAIILGKATTHEYAYGVTTP--
* ** . ** . ** ** ****:. .*:* * :*
MLBr_01702|M.leprae_Br4923 AYGPTRNPWNVDRVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALT
MAV_3859|M.avium_104 AYGPTRNPWNVERVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALT
Mb3036c|M.bovis_AF2122/97 AYGPTRNPWNLDRVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALT
Rv3011c|M.tuberculosis_H37Rv AYGPTRNPWNLDRVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALT
MMAR_1702|M.marinum_M AYGPTRNPWNLDRVPGGSGGGSAAALAAYQAPLAIGSDTGGSIRQPAALT
MUL_1939|M.ulcerans_Agy99 AYGPTRNPWNLDRVPGGSGGGSAAALAAYQAPLAIGSDTGGSIRQPAALT
Mflv_4246|M.gilvum_PYR-GCK AYGPTRNPWDTERVPGGSGGGSAAALAAFQAPLAIGTDTGGSIRQPAALT
Mvan_2115|M.vanbaalenii_PYR-1 AYGPTRNPWNVDCVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALT
MAB_3341c|M.abscessus_ATCC_199 AYGPTRNPWDVERVPGGSGGGSAAALAAFQAPLAIGTDTGGSIRQPAALT
TH_2639|M.thermoresistible__bu AFGPTRNPWDVNRVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALT
MSMEG_1088|M.smegmatis_MC2_155 ---PTRNPWSLDRIPGGSSGGTGAAIAAGVVAAGLGTDTGGSIRIPAALC
******. : :****.**:.**:** .. .:*:******* ****
MLBr_01702|M.leprae_Br4923 ATVGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTALLHAVIAGHDAR
MAV_3859|M.avium_104 ATVGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTALLHQVIAGHDIR
Mb3036c|M.bovis_AF2122/97 ATVGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTALLHQVIAGHDPR
Rv3011c|M.tuberculosis_H37Rv ATVGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTALLHQVIAGHDPR
MMAR_1702|M.marinum_M ATVGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTAMLHQVIAGHDAK
MUL_1939|M.ulcerans_Agy99 ATVGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTAMLHQVIAGHDAK
Mflv_4246|M.gilvum_PYR-GCK ATVGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTALLHQVIAGHDPL
Mvan_2115|M.vanbaalenii_PYR-1 ATVGVKPTYGTVSRYGLIACASSLDQGGPCARTVLDTALLHQVIAGHDPR
MAB_3341c|M.abscessus_ATCC_199 ATVGVKPTYGTVSRFGLVACASSLDQGGPCARTVLDTALLHQVIAGHDPR
TH_2639|M.thermoresistible__bu ATVGVKPTYGTVSRYGLVACASSLDQGGPCGRTVLDTALLHQVIAGHDAK
MSMEG_1088|M.smegmatis_MC2_155 GVTGHKPTFGLVSRSGVSALSSTLDHTGPLGRTVDDTVALLEVIAGHDPA
...* ***:* *** *: * :*:**: ** .*** **. * ******
MLBr_01702|M.leprae_Br4923 DSTSVEAAVPDIVGAAKAGESGDLHGVRVGVVRQLRG-EGYQSGVLASFQ
MAV_3859|M.avium_104 DSTSVDAPVPDVVGAARAGAAGDLKGVRVGVVKQLRG-EGYQPGVLASFE
Mb3036c|M.bovis_AF2122/97 DSTSVDAEVPDVVGAARAGAVGDLRGVRVGVVRQLHGGEGYQPGVLASFE
Rv3011c|M.tuberculosis_H37Rv DSTSVDAEVPDVVGAARAGAVGDLRGVRVGVVRQLHGGEGYQPGVLASFE
MMAR_1702|M.marinum_M DSTSLETEIPDVVGAAKAGASGDLRGVRIGVVKQLRG-DGYQPGVLASFE
MUL_1939|M.ulcerans_Agy99 DSTSLETEIPDVVGAAKAGASGDLRGVRIGVVKQLRG-DGYQPGVLASFE
Mflv_4246|M.gilvum_PYR-GCK DSTSVEAPVPDVVAAARTGAGGDLTGVRIGVVKQLRSGEGYQAGVLASFN
Mvan_2115|M.vanbaalenii_PYR-1 DSTSVNAAVPDVVGAARAGARGDLKGVRIGVVKQLRSGEGYQPGVLASFT
MAB_3341c|M.abscessus_ATCC_199 DSTSVDEPVPDVVAAARAGASGDLKGVRVGVVTQLR-GDGYQPGVMASFD
TH_2639|M.thermoresistible__bu DSTSLTAPVPDVVAAARAGASGDLKGVRVGVVRQLRG-EGIQPGVLASFD
MSMEG_1088|M.smegmatis_MC2_155 DPYSSSAPVPDFRAEFDRG----LAGLTIGVAEPYFC-DRLTPDVAAAFE
*. * :**. . * * *: :**. : ..* *:*
MLBr_01702|M.leprae_Br4923 AAVEQLIALGATVSEVDCPHFDYALAAYYLILPSEVSSNLARFDAMRYGL
MAV_3859|M.avium_104 AAVEQLTALGAEVSEVDCPHFEYALAAYYLILPSEVSSNLARFDAMRYGL
Mb3036c|M.bovis_AF2122/97 AAVEQLTALGAEVSEVDCPHFDHALAAYYLILPSEVSSNLARFDAMRYGL
Rv3011c|M.tuberculosis_H37Rv AAVEQLTALGAEVSEVDCPHFDHALAAYYLILPSEVSSNLARFDAMRYGL
MMAR_1702|M.marinum_M AAVAQLTALGAEVSEVDCPHFDHALAAYYLILPSEVSSNLARFDAMRYGL
MUL_1939|M.ulcerans_Agy99 AAVAQLTALGAEVSEVDCPHFDHALAAYYLILPSEVSSNLARFDAMRYGL
Mflv_4246|M.gilvum_PYR-GCK AAVDQLTALGAEVTEVDCPHFDYSLPAYYLILPSEVSSNLAKFDGMRYGL
Mvan_2115|M.vanbaalenii_PYR-1 AAVEQLTALGAEVSEVDCPHFDHSLAAYYLILPSEVSSNLAKFDGMRYGL
MAB_3341c|M.abscessus_ATCC_199 AAVEQLTALGADVVEVSCPHFEYALPAYYLILPSEVSSNLARFDAMRYGM
TH_2639|M.thermoresistible__bu AAVEQLAALGAEVIEVDCPNIDHALAAYYLIMPSEVSSNLARFDAMRYGL
MSMEG_1088|M.smegmatis_MC2_155 EAIHTLERGGATITSVTFTDVDLCPRVVDVVCGVEAAAWHAAQIGMAP--
*: * ** : .* ...: . . :: *.:: * .*
MLBr_01702|M.leprae_Br4923 RVGDDGSRSAEEVIALTRAAGFGPEVKRRIMIGAYALSAGYYDSYYSQAQ
MAV_3859|M.avium_104 RIGDDGSHSAEEVMALTRAAGFGPEVKRRIMIGTYALSAGYYDAYYNQAQ
Mb3036c|M.bovis_AF2122/97 RVGDDGTRSAEEVMAMTRAAGFGPEVKRRIMIGTYALSAGYYDAYYNQAQ
Rv3011c|M.tuberculosis_H37Rv RVGDDGTRSAEEVMAMTRAAGFGPEVKRRIMIGTYALSAGYYDAYYNQAQ
MMAR_1702|M.marinum_M RIGDDGTHSAEEVMAMTRAAGFGPEVKRRIMIGAYALSAGYYDAYYNQAQ
MUL_1939|M.ulcerans_Agy99 RIGDDGTHSAEEVMAMTRAAGFGPEVKRRIMIGAYALSAGYYDAYYNQAQ
Mflv_4246|M.gilvum_PYR-GCK RAGDDGTHSAEEVMALTRAAGFGPEVKRRIMIGAYALSAGYYDAYYNQAQ
Mvan_2115|M.vanbaalenii_PYR-1 RVGDDGTTSAEEVMAMTRAAGFGAEVKRRIMIGTYALSAGYYDAYYNQAQ
MAB_3341c|M.abscessus_ATCC_199 RVGDDGTHSAEEVMALTRAAGFGPEVKRRIMIGTYALSAGYYDAYYGRAQ
TH_2639|M.thermoresistible__bu RVGDDGTRSAEEVMALTRAAGFGPEVKRRIMIGTYALSAGYYDAYYNQAQ
MSMEG_1088|M.smegmatis_MC2_155 -------------------EKFSPEVR-----DALRLGASYTGVEYLEAR
*..**: .: *.*.* . * .*:
MLBr_01702|M.leprae_Br4923 KVRTLIARDLDKAYRS-VDVLVSPATPTTAFSLGEKADDP-----LAMYL
MAV_3859|M.avium_104 KVRTLIARDLDAAYES-VDVVVSPATPTTAFGLGEKVDDP-----LAMYL
Mb3036c|M.bovis_AF2122/97 KVRTLIARDLDAAYRS-VDVLVSPTTPTTAFRLGEKVDDP-----LAMYL
Rv3011c|M.tuberculosis_H37Rv KVRTLIARDLDAAYRS-VDVLVSPTTPTTAFRMGEKVDDP-----LAMYL
MMAR_1702|M.marinum_M KVRTLIARDLDEAYQS-VDVLVSPATPTTAFPLGEKVDDP-----LAMYL
MUL_1939|M.ulcerans_Agy99 KIRTLIARDLDEAYQS-VDVLVSPATPTTAFPLGEKVDDP-----LAMYL
Mflv_4246|M.gilvum_PYR-GCK KVRTLIARDLDAAYQK-VDVLVSPATPTTAFRLGEKVDDP-----LSMYL
Mvan_2115|M.vanbaalenii_PYR-1 KVRTLIARDLDEAYRS-VDVLVSPATPSTAFRLGEKVDDP-----LAMYL
MAB_3341c|M.abscessus_ATCC_199 KVRTLIKRDLERAYEQ-VDVLVTPATPTTAFRLGEKVDDP-----LAMYQ
TH_2639|M.thermoresistible__bu KVRTLVARDFDAAYEK-VDVLVSPTTPTTAFPLGEKVDHP-----LAMYL
MSMEG_1088|M.smegmatis_MC2_155 RARCAVIAGMNALFDSGPDLLISPTIAMTAPPYGSTHVDLGGRTVGVLDG
: * : .:: : . *::::*: . ** *.. . :
MLBr_01702|M.leprae_Br4923 FDLCTLPLNLAGHCGMSVPSGLSSDDDLPVGLQIMAPALADDRLYRVGAA
MAV_3859|M.avium_104 FDLCTLPLNLAGHCGMSVPSGLSPDDDLPVGLQIMAPALADDRLYRVGAA
Mb3036c|M.bovis_AF2122/97 FDLCTLPLNLAGHCGMSVPSGLSPDDGLPVGLQIMAPALADDRLYRVGAA
Rv3011c|M.tuberculosis_H37Rv FDLCTLPLNLAGHCGMSVPSGLSPDDGLPVGLQIMAPALADDRLYRVGAA
MMAR_1702|M.marinum_M FDLCTLPLNLAGHCGMSVPSGLSPDDGLPVGLQIMAPALADDRLYRVGAA
MUL_1939|M.ulcerans_Agy99 FDLCTLPLNLAGHCGMSVPSGLSPDDGLPVGLQIMAPALADDRLYRVGAA
Mflv_4246|M.gilvum_PYR-GCK FDLCTLPLNLAGHCGMSVPSGLSADDNLPVGLQIMAPALADDRLYRVGAA
Mvan_2115|M.vanbaalenii_PYR-1 FDLCTLPLNLAGHCGMSVPSGLSADDNLPVGLQIMAPALADDRLYRVGAA
MAB_3341c|M.abscessus_ATCC_199 FDLCTLPLNLAGHCGMSVPSGLSADDGLPVGLQIMAPALADDRLYRVGAA
TH_2639|M.thermoresistible__bu FDLCTLPLNLAGHCGMSVPSGLSPDDNLPVGLQIMAPALADDRLYRVGAA
MSMEG_1088|M.smegmatis_MC2_155 VNALTVPANVTGMPALTVPAGFGSD-GLPIGLQLMGRPGADATVLGAGRA
.: *:* *::* .::**:*:..* .**:***:*. . ** : .* *
MLBr_01702|M.leprae_Br4923 YEAARGPLPSAI--
MAV_3859|M.avium_104 YEAARGPLRSAI--
Mb3036c|M.bovis_AF2122/97 YEAARGPLLSAI--
Rv3011c|M.tuberculosis_H37Rv YEAARGPLLSAI--
MMAR_1702|M.marinum_M YEAARGPLPSAI--
MUL_1939|M.ulcerans_Agy99 YEAARGPLPSAI--
Mflv_4246|M.gilvum_PYR-GCK YEAARGPLPTAL--
Mvan_2115|M.vanbaalenii_PYR-1 YEAARGPLPTAL--
MAB_3341c|M.abscessus_ATCC_199 YETARGALPAAL--
TH_2639|M.thermoresistible__bu YEAARGPLPAAV--
MSMEG_1088|M.smegmatis_MC2_155 FERIAGFVDFTPNL
:* * : :