For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ADRRRVEDGIAESTLHLLRSGGPRAVTVEAVTSHSGIAKTTIYRRHHNRREMLAAALSGLATPESLDPGA ESTLHLLRSGGPRAVTVEAVTSHSGIAKTTIYRRHHNRREMLAAALSGLATPESLDPGADAPDRLRWVIA QAIGTIEDGIGIGGLAALCTEDDPEFNTLFRRILVEQRAALAEVIDAAKTDGSMRDDVDAQTLIDAVVGA YIAESARTGAVADGWQERLFALFWPIVRASA*
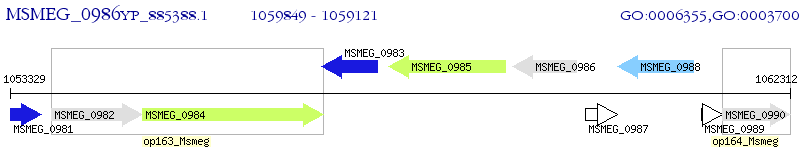
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0986 | - | - | 100% (242) | RemM protein |
| M. smegmatis MC2 155 | MSMEG_1430 | - | 3e-08 | 30.77% (169) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_3834 | - | 2e-07 | 27.17% (173) | putative TetR-family protein transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0437 | - | 1e-36 | 49.06% (159) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0832 | - | 9e-08 | 23.53% (170) | TetR family transcriptional regulator |
| M. marinum M | MMAR_4834 | - | 9e-06 | 23.90% (159) | TetR family transcriptional regulator |
| M. avium 104 | MAV_3805 | - | 6e-07 | 35.71% (70) | putative transcriptional regulator |
| M. thermoresistible (build 8) | TH_1099 | - | 3e-55 | 62.13% (169) | RemM protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0230 | - | 6e-39 | 44.95% (198) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0986|M.smegmatis_MC2_155 --------------MADRR--RVEDGIAESTLHLLRSGGPRAVTVEAVTS
TH_1099|M.thermoresistible__bu --------------MAVR---DVDDGIADATLALLRTKGPRSATVQAVAA
Mflv_0437|M.gilvum_PYR-GCK --------------MAANSGDDVGARILDAALELLRARGPKAVTMQAVVD
Mvan_0230|M.vanbaalenii_PYR-1 ----MRRQQHRRRQMAAQSTDEVGARILDAALELLRARGPRAVTMQAIVD
MMAR_4834|M.marinum_M MERDTLEFPVDDDDDVDPRRLRSRTRLLDAATKLLSAGGIEAVTIDAVTK
MAV_3805|M.avium_104 MGTQPAETP-DDEDDVDPRRTRSRTRLLDAAANLLKTGGIEAVTIDAVTK
MAB_0832|M.abscessus_ATCC_1997 ---------MDKRQGGRPRDPDKDDAVRETVRRMLAADGYQRTTIPAVAR
: ::. :* : * . .*: *:.
MSMEG_0986|M.smegmatis_MC2_155 HSGIAKTTIYRRHHNRREMLAAALSGLATPESLDPGAESTLHLLRSGGPR
TH_1099|M.thermoresistible__bu QSGIAKTTIYRRFRDRRDMLSAALSRVAHPDPLEP---------------
Mflv_0437|M.gilvum_PYR-GCK ATGIAKTTIYRRHPNRRTLLAAALETLGERPSVPA---------------
Mvan_0230|M.vanbaalenii_PYR-1 ATGIAKTTIYRRHQNRRMLLTAALETLGERPSAPP---------------
MMAR_4834|M.marinum_M ASKVARTTLYRHFSSSTQLLAATFERLLPQVHPP----------------
MAV_3805|M.avium_104 ASKVARTTLYRHFNSSSQLLAATFERLLPQVIPPA---------------
MAB_0832|M.abscessus_ATCC_1997 AAGIGAPTIYRRWPTQAAMVESAIADRPWPEELAA---------------
: :. .*:**: :: :::
MSMEG_0986|M.smegmatis_MC2_155 AVTVEAVTSHSGIAKTTIYRRHHNRREMLAAALSGLATPESLDPGADAPD
TH_1099|M.thermoresistible__bu --------------------------------------------DTAAPD
Mflv_0437|M.gilvum_PYR-GCK --------------------------------------------DATRHE
Mvan_0230|M.vanbaalenii_PYR-1 --------------------------------------------ESGRAE
MMAR_4834|M.marinum_M ------------------------------------------PATGSMRD
MAV_3805|M.avium_104 ------------------------------------------PTSGTLRE
MAB_0832|M.abscessus_ATCC_1997 --------------------------------------------PSEFRY
MSMEG_0986|M.smegmatis_MC2_155 RLRWVIAQAIGTIEDG---IGIGGLAALCTEDDPEFNTLFRR--------
TH_1099|M.thermoresistible__bu RLRWLIAQAVKTIEVG---IGYGGFAAMLTDEDPDFTAVFRE--------
Mflv_0437|M.gilvum_PYR-GCK RLNWVITQSVDVIAHG---IGAGGFAALLTDDDPEFSDAFRA--------
Mvan_0230|M.vanbaalenii_PYR-1 RLQWVITQSVDVIAAG---IGAGGFAALLTDEDPEFSEAFRA--------
MMAR_4834|M.marinum_M QLIELLSRQATLFQEAPLHVTTLAWVALGPTPDGTQETQDRH---ALRAR
MAV_3805|M.avium_104 RLIELLSRQADLFAEAPVQVTTLAWAALGPAETHEPNTVQHAGAGTLRMR
MAB_0832|M.abscessus_ATCC_1997 YLPVLVRAVVGYFADPATRAAMPGLLVEYYREPSQYVVLAER--------
* :: : . . .
MSMEG_0986|M.smegmatis_MC2_155 ILVEQRAALAEVIDAAKTDGSMRDDVDAQTLIDAVVGAYIAESARTGAVA
TH_1099|M.thermoresistible__bu ILAGQRAELEAVIDGGKADGSIRADVDGSALIDAVVGTHIAERARVGEVA
Mflv_0437|M.gilvum_PYR-GCK ILGAYRRHAIDALDVDVATG--------ETVIDLIVGGYVAELARTGSVD
Mvan_0230|M.vanbaalenii_PYR-1 ILVAYRRQAIDALAIGDRVG--------DTLVDIIVGSYVAELARTGSID
MMAR_4834|M.marinum_M IIDQYRQPFVALLQSPEARADLDDFDMELILCQLIGPVAFARLTGLRAID
MAV_3805|M.avium_104 VIEQYRRPFDEILYSPESVDQLGELDIELALCQLVGPLVFARMTGLRVIT
MAB_0832|M.abscessus_ATCC_1997 TEMPLREAFRRAHAGAVGSGERADQPGADALFDTIVGMAVYHGVLRGTAD
* : : : . . .
MSMEG_0986|M.smegmatis_MC2_155 DGWQERLFALFWPIVRASA--------------------
TH_1099|M.thermoresistible__bu ADWENRLFDLFWPTVRP----------------------
Mflv_0437|M.gilvum_PYR-GCK DGWNARMADAVERVLPSR---------------------
Mvan_0230|M.vanbaalenii_PYR-1 DGWAARIAAVLDGVLDAAAND------------------
MMAR_4834|M.marinum_M RQDCERIVDDFLVAHRRQADKPAQRAVSRKPVSRRSGSD
MAV_3805|M.avium_104 HQDCTRIVEGFIAAQT--GDQPAWVEAS-SPNQ------
MAB_0832|M.abscessus_ATCC_1997 DALVEQILDVVDAASSVGRGLHLDETNSDKERKR-----
:: .