For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTVFSRPGAADARMSFESRYDNFIGGEWVAPVDGRYFENPTPVTGQVFCEVARSSSADIEKALDAAHAAA PAWGKTAPAERALILNRIADRMEQNLEALALAEAWDNGKPIRETLNADIPLAIDHFRYFASAIRAQEGSL SQVDEDTVAYHFHEPLGVVGQIIPWNFPILMAVWKLAPALAAGNAVVLKPAEQTPASVLYLMSLIADLVP AGVVNIVNGFGVEAGKPLASSNRIAKIAFTGETTTGRLIMQYASQNLIPVTLELGGKSPNIFFSDVMAAA DDFQDKALEGFTMFALNQGEVCTCPSRSLIQSDIFDEFLELAAIRTKAVRQGDPLDTETMIGSQASNDQF EKILSYIEIGKSEGARIVTGGERADLGGDLSGGYYIQPTIFTGHNKMRIFQEEIFGPVVAVTSFSDYDDA ISIANDTLYGLGAGVWSRDGNTAYRAGRDIKAGRVWTNCYHQYPAHAAFGGYKQSGIGRENHKMMLDHYQ QTKNLLVSYSDKAQGFF
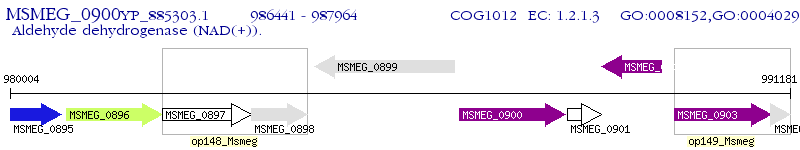
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0900 | - | - | 100% (507) | eptc-inducible aldehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1543 | - | 0.0 | 91.91% (507) | eptc-inducible aldehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1306 | - | 0.0 | 70.41% (507) | aldehyde dehydrogenase (NAD) family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0467 | - | 0.0 | 86.59% (507) | aldehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4952 | - | 0.0 | 88.17% (507) | aldehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv0458 | - | 0.0 | 86.59% (507) | aldehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 1e-50 | 31.11% (450) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2792c | - | 0.0 | 84.42% (507) | aldehyde dehydrogenase |
| M. marinum M | MMAR_4912 | - | 0.0 | 88.36% (507) | aldehyde dehydrogenase |
| M. avium 104 | MAV_4691 | - | 0.0 | 88.36% (507) | eptc-inducible aldehyde dehydrogenase |
| M. thermoresistible (build 8) | TH_3284 | - | 0.0 | 91.09% (494) | PUTATIVE eptc-inducible aldehyde dehydrogenase |
| M. ulcerans Agy99 | MUL_0494 | - | 0.0 | 87.77% (507) | aldehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1468 | - | 0.0 | 89.15% (507) | aldehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0467|M.bovis_AF2122/97 MTVFSRPGSAGALMSYESRYQNFIGGQWVAPVHGRYFENPTPVTGQPFCE
Rv0458|M.tuberculosis_H37Rv MTVFSRPGSAGALMSYESRYQNFIGGQWVAPVHGRYFENPTPVTGQPFCE
MAV_4691|M.avium_104 MTVFARPGSAGALMSYESRYDNFIGGQWVAPARGRYFENPTPVTGQTFCE
Mflv_4952|M.gilvum_PYR-GCK MTVYARPGADGSLMSFKPRYDNFIGGQWVAPTAGRYFENPSPVTGQPFTE
Mvan_1468|M.vanbaalenii_PYR-1 MTVYARPGADGSLMSFKPRYDNFIGGQWVAPNAGRYFENPSPVTGQPFTE
TH_3284|M.thermoresistible__bu -------------MSFESRYDNFIGGEWVAPVGGEYFENPSPVTGQPFCE
MSMEG_0900|M.smegmatis_MC2_155 MTVFSRPGAADARMSFESRYDNFIGGEWVAPVDGRYFENPTPVTGQVFCE
MMAR_4912|M.marinum_M MTVFARPGAHSSLMTFESRYGNFIGGEWVAPAAGEYFEDLTPVTGAPFCE
MUL_0494|M.ulcerans_Agy99 MTVFARPGVHSSLMTFESRYDNFIGGEWVAPAAGEYFEDLTPVTGAPFCE
MAB_2792c|M.abscessus_ATCC_199 MTVYARPGAQGSLMTFASRYENFIGGQWVPPEHGRYFENRTPVTGEVFCE
MLBr_02573|M.leprae_Br4923 ----------------------------------MSIATINPATGETVKT
: .*.** .
Mb0467|M.bovis_AF2122/97 VPRSDAADIDKALDAAHAAAPGWGKTAPAERAAILNMIADRIDKNAAALA
Rv0458|M.tuberculosis_H37Rv VPRSDAADIDKALDAAHAAAPGWGKTAPAERAAILNMIADRIDKNAAALA
MAV_4691|M.avium_104 VARSDEADVDKALDAAHAAAPAWGKTAPPERAAILNKIADRIEENLESLA
Mflv_4952|M.gilvum_PYR-GCK VARSDEADIEKALDAAHAAAPAWGKTSAAERAVILNKIADRIEENLESIA
Mvan_1468|M.vanbaalenii_PYR-1 VARSDESDIEKALDAAHAAAPAWGKTSAAERAVILNKIADRIEENLESIA
TH_3284|M.thermoresistible__bu VARSTEADIDKALDAAHAAAPAWGKTAPAERALILNRIADRIEANLESIA
MSMEG_0900|M.smegmatis_MC2_155 VARSSSADIEKALDAAHAAAPAWGKTAPAERALILNRIADRMEQNLEALA
MMAR_4912|M.marinum_M VPRSTDVDVEKALDAAHAAAPAWGKAAPAERAAILHKIADRMEANLEALA
MUL_0494|M.ulcerans_Agy99 VPRSTDVDVEKALDAAHAAAPAWGKAAPAERATILHKIADRMEANLEALA
MAB_2792c|M.abscessus_ATCC_199 IPRSDEADVEKALDAAHAAAPGWAKLSPADRALILNRVADRMEANLEAIA
MLBr_02573|M.leprae_Br4923 FTPTTQDEVEDAIARAYARFDNYRHTTFTQRAKWANATADLLEAEANQSA
.. : :::.*: *:* : : : .:** : ** :: : *
Mb0467|M.bovis_AF2122/97 VAEVWDNGKPVREALAADIPLAVDHFRYFAA---AIRAQEGALSQIDEDT
Rv0458|M.tuberculosis_H37Rv VAEVWDNGKPVREALAADIPLAVDHFRYFAA---AIRAQEGALSQIDEDT
MAV_4691|M.avium_104 VAEVWDNGKPIREALAADIPLAADHFRYFAG---ALRAQEGSLSQIDDDT
Mflv_4952|M.gilvum_PYR-GCK LAESWDNGKPIRETLNADLPLAVDHFRYFAG---CIRAQEGSLSEVDADT
Mvan_1468|M.vanbaalenii_PYR-1 LAESWDNGKPIRETLNADIPLAVDHFRYFAG---AIRAQEGSLSEIDDDT
TH_3284|M.thermoresistible__bu LAESWDNGKPIRETLNADIPLAVDHFRYFAG---AIRAQEGSLSQIDDDT
MSMEG_0900|M.smegmatis_MC2_155 LAEAWDNGKPIRETLNADIPLAIDHFRYFAS---AIRAQEGSLSQVDEDT
MMAR_4912|M.marinum_M LAESWDNGKPIRETLNADIPLAIDHFRYFAG---AIRAQEGSLSQIDSDT
MUL_0494|M.ulcerans_Agy99 LAESWDNGKPIREALNADIPLAIDHFRYFAG---AIRAQEGSLSQIDSDT
MAB_2792c|M.abscessus_ATCC_199 VAESWDNGKPIRETVNADVPLAIDHFRYFAG---VLRAQEGSLSQIDEDT
MLBr_02573|M.leprae_Br4923 ALMTLEMGKTLQSAKDEAMKCAKG-FRYYAEKAETLLADEPAEAAAVGAS
: **.::.: : * . ***:* : *:* : : :
Mb0467|M.bovis_AF2122/97 VAYHFHEPLGVVGQIIPWNFPILMAAWKLAPALAAGNTAVLKPAEQTPAS
Rv0458|M.tuberculosis_H37Rv VAYHFHEPLGVVGQIIPWNFPILMAAWKLAPALAAGNTAVLKPAEQTPAS
MAV_4691|M.avium_104 VAYHFHEPLGVVGQIIPWNFPILMGAWKLAPALAAGNAVVLKPAEQTPVS
Mflv_4952|M.gilvum_PYR-GCK VAYHFHEPLGVVGQIIPWNFPILMAVWKLAPALAAGNAVVLKPAEQTPAS
Mvan_1468|M.vanbaalenii_PYR-1 VAYHFHEPLGVVGQIIPWNFPILMAVWKLAPALAAGNAIVLKPAEQTPAS
TH_3284|M.thermoresistible__bu VAYHFHEPLGVVGQIIPWNFPILMATWKLAPALAAGNAVVLKPAEQTPAS
MSMEG_0900|M.smegmatis_MC2_155 VAYHFHEPLGVVGQIIPWNFPILMAVWKLAPALAAGNAVVLKPAEQTPAS
MMAR_4912|M.marinum_M VAYHFHEPLGVVGQIIPWNFPLLMATWKLAPALAAGNAVVLKPAEQTPAS
MUL_0494|M.ulcerans_Agy99 VAYHFHEPLGVVGQIIPWNFPLLMATWKLAPALAAGNAVVLKPAEQTPAS
MAB_2792c|M.abscessus_ATCC_199 VAYHFHEPLGVVGQIIPWNFPLLMASWKLAPALAAGNTVVLKPAEQTPAS
MLBr_02573|M.leprae_Br4923 KALARYQPLGVVLAVMPWNFPLWQTVRFAAPALMAGNVGLLKHASNVPQC
* ::***** ::*****: **** ***. :** *.:.* .
Mb0467|M.bovis_AF2122/97 VLYLMSLIGDLLPPGVVNVVNGFGAEAGKPLASSDRIAKVAFTGETTTGR
Rv0458|M.tuberculosis_H37Rv VLYLMSLIGDLLPPGVVNVVNGFGAEAGKPLASSDRIAKVAFTGETTTGR
MAV_4691|M.avium_104 VLYLMSLIGDLLPPGVVNVVNGFGAEAGKPLASSNRIAKIAFTGETTTGR
Mflv_4952|M.gilvum_PYR-GCK ILYLMEVIGDLLPAGVVNVVNGFGVEAGKPLASSNRIAKIAFTGETTTGR
Mvan_1468|M.vanbaalenii_PYR-1 ILYLMELIADLLPAGVINVVNGFGVEAGKPLASSNRIAKIAFTGETTTGR
TH_3284|M.thermoresistible__bu VMYLMSLIGDLLPPGVVNVVNGFGVEAGKPLASSNRIAKIAFTGETTTGR
MSMEG_0900|M.smegmatis_MC2_155 VLYLMSLIADLVPAGVVNIVNGFGVEAGKPLASSNRIAKIAFTGETTTGR
MMAR_4912|M.marinum_M ILYLMSLIGDLLPAGVVNVVNGFGVEAGKPLASSNRIAKIAFTGETTTGR
MUL_0494|M.ulcerans_Agy99 ILYLMSLIGDLLPAGVVNVVNGFGVEAGKPLASSNRIAKIAFTGETTTGR
MAB_2792c|M.abscessus_ATCC_199 ILHLFSLIGDLLPPGVVNIVNGFGVEAGKPLASSSRIAKIAFTGETSTGR
MLBr_02573|M.leprae_Br4923 ALYLADVIARGGFPEDCFQTLLISANGVEAILRDPRVAAATLTGSEPAGQ
::* .:*. . . :..:. :.: . *:* ::**. .:*:
Mb0467|M.bovis_AF2122/97 LIMQYASHNLIPVTLELGGKSPNIFFADVLAAHDDFCDKALEGFTMFALN
Rv0458|M.tuberculosis_H37Rv LIMQYASHNLIPVTLELGGKSPNIFFADVLAAHDDFCDKALEGFTMFALN
MAV_4691|M.avium_104 LIMQYASQNLIPVTLELGGKSPNIFFSDVMAKADDYQDKALEGFTMFALN
Mflv_4952|M.gilvum_PYR-GCK LIMQYASQNLIPVTLELGGKSPNIFFNDVLAASDDYQDKALEGFTMFALN
Mvan_1468|M.vanbaalenii_PYR-1 LIMQYASQNLIPVTLELGGKSPNIFFNDVLAQADNYQDKALEGFTMFALN
TH_3284|M.thermoresistible__bu LIMQYASQNLIPVTLELGGKSPNIFFADVMAASDDFCDKALEGFTMFALN
MSMEG_0900|M.smegmatis_MC2_155 LIMQYASQNLIPVTLELGGKSPNIFFSDVMAAADDFQDKALEGFTMFALN
MMAR_4912|M.marinum_M LIMQYASQNLIPVTLELGGKSPNIFFSDVLAAHDDYQDKALEGFTMFALN
MUL_0494|M.ulcerans_Agy99 LIMQYASQNLIPVTLELGGKSPNIFFPDVLAAHDDYQDKALEGFTMFALN
MAB_2792c|M.abscessus_ATCC_199 LIMQYASENLIPVTLELGGKSPNLFFSDVMAGGDDFQDKALEGFTMFAFN
MLBr_02573|M.leprae_Br4923 AVGAIAGNEIKPTVLELGGSDP-----FIVMPSADLDAAVATAVTGRVQN
: *..:: *..*****..* :: : . ..* . *
Mb0467|M.bovis_AF2122/97 QGEVCTCPSRSLIQADIYDEFLELAAIRTKAVRQGDPLDTETMLGSQASN
Rv0458|M.tuberculosis_H37Rv QGEVCTCPSRSLIQADIYDEFLELAAIRTKAVRQGDPLDTETMLGSQASN
MAV_4691|M.avium_104 QGEVCTCPSRSLIQADIYDEFLELAAIRTKAVRQGDPLDSETMLGSQASN
Mflv_4952|M.gilvum_PYR-GCK QGEVCTCPSRSLIQADIYDEFLALAAIRTKAVRQGDPLDTETMIGAQASN
Mvan_1468|M.vanbaalenii_PYR-1 QGEVCTCPSRSLIQADIYDEFLALAAIRTKAVRQGDPLDTETMIGAQASN
TH_3284|M.thermoresistible__bu QGEVCTCPSRSLIQADIYDEFLELATIRTKAVRQGDPLDTETMVGAQASN
MSMEG_0900|M.smegmatis_MC2_155 QGEVCTCPSRSLIQSDIFDEFLELAAIRTKAVRQGDPLDTETMIGSQASN
MMAR_4912|M.marinum_M QGEVCTCPSRGLIQADIYDRFLELAAIRTKAVRQGDPLDTETMIGAQASN
MUL_0494|M.ulcerans_Agy99 QGEVCTCPSRGLIQADIYDRFLELAAIRTKAVRQGDPLDTETMIGAQASN
MAB_2792c|M.abscessus_ATCC_199 QGEVCTCPSRSLIQADIFDDFLELAAIRTKAIRQGDPLDTETMMGAQASH
MLBr_02573|M.leprae_Br4923 NGQSCIAAKRFIAHADIYDEFVEKFVARMETLRVGDPTDPDTDVGPLATE
:*: * ...* : ::**:* *: . * :::* *** *.:* :*. *:.
Mb0467|M.bovis_AF2122/97 DQLEKVLSYIEIGKQEGAVIIAGGERAELGGDLSGGYYMQPTIFTGTNNM
Rv0458|M.tuberculosis_H37Rv DQLEKVLSYIEIGKQEGAVIIAGGERAELGGDLSGGYYMQPTIFTGTNNM
MAV_4691|M.avium_104 DQLEKVLSYIAIGKDEGATIITGGERAELGGDLSGGFYMQPTIFAGNNKM
Mflv_4952|M.gilvum_PYR-GCK DQLEKILSYIEIGKSEGAKVVTGGERAELGGDLNGGYYVAPTIFTGNNTM
Mvan_1468|M.vanbaalenii_PYR-1 DQLEKILSYIEIGKSEGAQVVTGGERAELGGDLNGGYYVAPTIFTGHNKM
TH_3284|M.thermoresistible__bu DQLEKILSYIDIGRSEGATVVTGGERAELGGDLSGGYYVQPTIFTGDNSM
MSMEG_0900|M.smegmatis_MC2_155 DQFEKILSYIEIGKSEGARIVTGGERADLGGDLSGGYYIQPTIFTGHNKM
MMAR_4912|M.marinum_M DQLQKILSYIEIGKEEGARVVTGGERCELGGDLNGGYYVQPTIFEGTNNM
MUL_0494|M.ulcerans_Agy99 DQLQKILSYIEIGKEEGARVVTGGERCELGGDLNGGYYVQPTIFEGTNNM
MAB_2792c|M.abscessus_ATCC_199 EQLEKILSYIDIGKSEGARLVTGGDRADLGGDLSGGYYVTPTVFAGDNSM
MLBr_02573|M.leprae_Br4923 SGRAQVEQQVDAAAAAGAVIRCGGKRPDR----PGWFYPPTVITDITKDM
. :: . : . ** : **.* : * :* ..: : *
Mb0467|M.bovis_AF2122/97 RIFKEEIFGPVVAVTSFTDYDDAIGIANDTLYGLGAGVWSRDGNTAYRAG
Rv0458|M.tuberculosis_H37Rv RIFKEEIFGPVVAVTSFTDYDDAIGIANDTLYGLGAGVWSRDGNTAYRAG
MAV_4691|M.avium_104 RIFQEEIFGPVVAVTSFTDYDDAISIANDTLYGLGAGVWSRDGNTAYRAG
Mflv_4952|M.gilvum_PYR-GCK RIFQEEIFGPVVAVTSFTDYDDAISLANDTLYGLGAGVWSRNGNTAYRAG
Mvan_1468|M.vanbaalenii_PYR-1 RIFQEEIFGPVVAVTSFTDYDDAISIANDTLYGLGAGVWSRDGNTAYRAG
TH_3284|M.thermoresistible__bu RIFQEEIFGPVVAVTSFTDYDDAIRIANDTLYGLGAGVWSRDGNTAYRAG
MSMEG_0900|M.smegmatis_MC2_155 RIFQEEIFGPVVAVTSFSDYDDAISIANDTLYGLGAGVWSRDGNTAYRAG
MMAR_4912|M.marinum_M RIFQEEIFGPVVSVTSFTDYDDAIRIANDTLYGLGAGVWSRDGNTAYRAG
MUL_0494|M.ulcerans_Agy99 RIFQEEIFGPVVSVTLFTDYDDAIRIANDTLYGLGAGVWSRDGNTAYRAG
MAB_2792c|M.abscessus_ATCC_199 RIFQEEIFGPVLAVTSFADYDDAIAIANDTPYGLGAGVWSRNGNIAYRAG
MLBr_02573|M.leprae_Br4923 ALYTDEVFGPVASVYCATNIDDAIEIANATPFGLGSNAWTQNESEQRHFI
:: :*:**** :* :: **** :** * :***:..*::: . :
Mb0467|M.bovis_AF2122/97 RDIQAGRVWVNCYHLYPAHAAFGGYKQSGIGREGHQMMLQHYQHTKNLLV
Rv0458|M.tuberculosis_H37Rv RDIQAGRVWVNCYHLYPAHAAFGGYKQSGIGREGHQMMLQHYQHTKNLLV
MAV_4691|M.avium_104 RDIQAGRVWVNCYHAYPAHAAFGGYKQSGIGRENHKMMLDHYQQTKNMLV
Mflv_4952|M.gilvum_PYR-GCK RDIKAGRVWTNCYHQYPAHAAFGGYKQSGIGRENHRMMLDHYQQTKNLLV
Mvan_1468|M.vanbaalenii_PYR-1 RDIKAGRVWTNCYHQYPAHAAFGGYKQSGIGRENHKMMLDHYQQTKNLLV
TH_3284|M.thermoresistible__bu RDIKAGRVWTNCYHVYPAHAAFGGYKQSGIGRENHTMMLDHYQQTKNLLV
MSMEG_0900|M.smegmatis_MC2_155 RDIKAGRVWTNCYHQYPAHAAFGGYKQSGIGRENHKMMLDHYQQTKNLLV
MMAR_4912|M.marinum_M RDIKAGRVWTNCYHAYPAHAAFGGYKQSGIGRENHKMMLDHYQQTKNLLV
MUL_0494|M.ulcerans_Agy99 RDIKAGRVWTNCYHAYPAHAAFGGYKQSGIGRENHKMMLDHYQQTKNLLV
MAB_2792c|M.abscessus_ATCC_199 RDIKAGRVWTNCYHQYPAHAAFGGYKRSGIGRENHKMMLDHYQQTKNLLV
MLBr_02573|M.leprae_Br4923 DNIVAGQVFINGMTVSYPELPFGGIKRSGYGRELSTHGIREFCNIKTVWI
:* **:*: * .. .*** *:** *** : .: : *.: :
Mb0467|M.bovis_AF2122/97 SYSDKALGFF
Rv0458|M.tuberculosis_H37Rv SYSDKALGFF
MAV_4691|M.avium_104 SYSNKAQGFF
Mflv_4952|M.gilvum_PYR-GCK SYSDQALGFF
Mvan_1468|M.vanbaalenii_PYR-1 SYSDKALGFF
TH_3284|M.thermoresistible__bu SYSTKAQGFF
MSMEG_0900|M.smegmatis_MC2_155 SYSDKAQGFF
MMAR_4912|M.marinum_M SYGSKAQGFF
MUL_0494|M.ulcerans_Agy99 SYGSKAQGFF
MAB_2792c|M.abscessus_ATCC_199 SYGSKAQGFF
MLBr_02573|M.leprae_Br4923 A---------
: