For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTKTDQQEAAELRDTVHPESQTSDKQPSTGMQKRVLAGGGIGQFIEFYDFTLYGLSAVTLAKLFFPGGSS LAGLLATFATFGVAFFVRPLGGLFFGALGDRIGRRKVLFLTLLLIGTATALIGMLPTHASIGIAAPILLV TCRLVQGFSAGGESVGAPAFVFEHAPLERRGFWLNITISATALPSVVGGTLILILSSTMPAAAFESYGWR IPFLLALPLAVFGVWIRAKTEESEAFKQVLAKHEHKEFSPIRETFRQNKVRMAQVIFVMGLTAMGFYFLS GYFVSYVQTTGNLSREQSLLINAIAMAIYALLLPVGGIIGDRVGRKPMLIAGSAALAVLSIPCFMLVTSG SLSLAMVGQLLFVVPLCIYGGGCYTFFVEVFTTATRFTSAAISYNVAYAVFGGTAPFIGTMLVSSTELDA APGYYMATAAAVVLLLLLFARVPETRRRIG
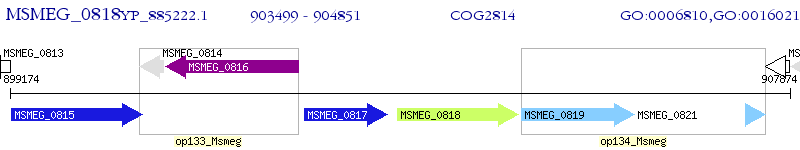
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0818 | - | - | 100% (450) | transporter, major facilitator family protein |
| M. smegmatis MC2 155 | MSMEG_4889 | - | 2e-82 | 38.82% (407) | integral membrane transport protein |
| M. smegmatis MC2 155 | MSMEG_1477 | - | 2e-72 | 35.82% (455) | major facilitator superfamily protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1232 | - | 2e-53 | 32.95% (434) | integral membrane transport protein |
| M. gilvum PYR-GCK | Mflv_1075 | - | 5e-86 | 40.62% (416) | general substrate transporter |
| M. tuberculosis H37Rv | Rv1200 | - | 2e-53 | 32.95% (434) | integral membrane transport protein |
| M. leprae Br4923 | MLBr_01562 | - | 5e-10 | 22.65% (309) | putative transmembrane efflux protein |
| M. abscessus ATCC 19977 | MAB_2263c | - | 4e-65 | 34.79% (434) | major facilitator transporter |
| M. marinum M | MMAR_4238 | - | 6e-53 | 32.08% (424) | integral membrane transport protein |
| M. avium 104 | MAV_4627 | - | 7e-67 | 37.73% (432) | major facilitator superfamily protein |
| M. thermoresistible (build 8) | TH_3722 | - | 9e-79 | 36.87% (434) | PUTATIVE General substrate transporter |
| M. ulcerans Agy99 | MUL_0949 | - | 2e-52 | 31.84% (424) | integral membrane transport protein |
| M. vanbaalenii PYR-1 | Mvan_5748 | - | 7e-85 | 40.34% (409) | general substrate transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1075|M.gilvum_PYR-GCK ------------------------------MEVQRD--------SDIRRV
Mvan_5748|M.vanbaalenii_PYR-1 ----------------------------------ME--------TDIRRV
TH_3722|M.thermoresistible__bu --------------------VSENPSIVSAPPAPDA--------VVIRKA
MAB_2263c|M.abscessus_ATCC_199 -----------------MSSAGPREGETEPPPVPRPDFQSPEGRRQLHRA
MSMEG_0818|M.smegmatis_MC2_155 ---------------MTKTDQQEAAELRDTVHPESQTSDKQPSTGMQKRV
Mb1232|M.bovis_AF2122/97 ----------------------------------------------MKRV
Rv1200|M.tuberculosis_H37Rv ----------------------------------------------MKRV
MMAR_4238|M.marinum_M ----------------------------------------------MSRV
MUL_0949|M.ulcerans_Agy99 ----------------------------------------------MSRV
MAV_4627|M.avium_104 -----------------MAATLPAPAPAGLADGHPLDESAEQQRRRLRIV
MLBr_01562|M.leprae_Br4923 MVTLYDHERAVHNWTAARSDRPAPSRLTQQVEPASERTSKYPTLLPSGRF
Mflv_1075|M.gilvum_PYR-GCK VTGASIGNAVEWFDFAIYG-FLATFIAAQFFPAGNDTAALLNTFAIFAAA
Mvan_5748|M.vanbaalenii_PYR-1 VTGASIGNAVEWFDFAIYG-FLATFIAAHFFPAGNDTAALLNTFAIFAAA
TH_3722|M.thermoresistible__bu VTGASIGNAVEWFDFAIYG-FLATYIAANFFPAGDDTAALLNTFAIFAAA
MAB_2263c|M.abscessus_ATCC_199 IAASAMGNATEWYDYGVYA-ATTTFLTQAFFPTLGTAFTMLG----YAIS
MSMEG_0818|M.smegmatis_MC2_155 LAGGGIGQFIEFYDFTLYG-LSAVTLAKLFFPGGSSLAGLLATFATFGVA
Mb1232|M.bovis_AF2122/97 ALACLVGSAIEFYDFLIYGTAAALVFPTVFFPHLDPTVAAVASMGTFAVA
Rv1200|M.tuberculosis_H37Rv ALACLVGSAIEFYDFLIYGTAAALVFPTVFFPHLDPTVAAVASMGTFAVA
MMAR_4238|M.marinum_M ALACVVGSTVEYYDFCIYGTAAALVFPAVFYPGLSPALATIASMGTFATA
MUL_0949|M.ulcerans_Agy99 ALACVVGSTVEYYDFCIYGTAAALVFPAVFYPGLSPALATIASMGTFATA
MAV_4627|M.avium_104 VAASLLGTTVEWYDFFLYATAAGLVFNKVFFPNESSLVGTLLAFATFAVG
MLBr_01562|M.leprae_Br4923 PSGRFIAAVIVIGGMQLLA-TMDSTVAIVALPKIQNELSLSDAGRSWGIT
. :. . : . . * :.
Mflv_1075|M.gilvum_PYR-GCK FFMRPLGGFFFG--PLGDRIGRQKVLAVVILLMSAATLCIGLLPTYDAIG
Mvan_5748|M.vanbaalenii_PYR-1 FFMRPLGGFFFG--PLGDRIGRQKVLAVVILLMSAATLGIGLLPTYEAIG
TH_3722|M.thermoresistible__bu FVVRPLGGFFFG--PLGDRIGRQRVLALVILLMSGATLTMGLLPTYESIG
MAB_2263c|M.abscessus_ATCC_199 FLLRPLGGMVWG--PLGDRLGRKSVMAITIVLMALSTTLIGILPSWATIG
MSMEG_0818|M.smegmatis_MC2_155 FFVRPLGGLFFG--ALGDRIGRRKVLFLTLLLIGTATALIGMLPTHASIG
Mb1232|M.bovis_AF2122/97 FLSRPFGAAVFG--YFGDRLGRKKTLVATLLIMGLATVTVGLVPTTVAIG
Rv1200|M.tuberculosis_H37Rv FLSRPFGAAVFG--YFGDRLGRKKTLVATLLIMGLATVTVGLVPTTVAIG
MMAR_4238|M.marinum_M FLSRPLGAAVFG--HFGDRLGRKKTLVATLLIMALSTVTVGVVPSTATIG
MUL_0949|M.ulcerans_Agy99 FLSRPLGAAVFG--HFGDRLGRKKTLVATLLIMALSTVTVGVVPSTATIG
MAV_4627|M.avium_104 FVMRPIGGLVFG--HIGDRIGRKRSLALTMLIMGGATALIGVLPTAAQIG
MLBr_01562|M.leprae_Br4923 AYVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLCAVAWDEVTLV
.:*. . :** :**: : : :: :: .: :
Mflv_1075|M.gilvum_PYR-GCK VAAPLLLLVLRCLQGFSAGGEYGGGAVYLAEFASDRRRGLTITFMAWSGV
Mvan_5748|M.vanbaalenii_PYR-1 VAAPMLLLVLRCLQGFSAGGEYGGGAVYLAEFASDARRGLTITFMAWSGV
TH_3722|M.thermoresistible__bu VLAPLLLLLLRCLQGFSAGGEYGGGAVYLAEFAPDRRRGLIVTFMAWSGV
MAB_2263c|M.abscessus_ATCC_199 MAAPALLILLRVIQGFSTGGEYGGAATFMAEFAPDKKRGKYGSFLEFGTL
MSMEG_0818|M.smegmatis_MC2_155 IAAPILLVTCRLVQGFSAGGESVGAPAFVFEHAPLERRGFWLNITISATA
Mb1232|M.bovis_AF2122/97 AAAPLILTTMRLLQGFAVGGEWAGSALLSAEYAPASKRGWYGMFTVVGGG
Rv1200|M.tuberculosis_H37Rv AAAPLILTTMRLLQGFAVGGEWAGSALLSAEYAPASKRGWYGMFTVVGGG
MMAR_4238|M.marinum_M VSAPLILVALRLLQGFAVGGEWAGSALLSIESAPAHKRGYYGMFTVLGGG
MUL_0949|M.ulcerans_Agy99 VSAPLILVALRLLQGFAVGGEWAGSALLSIESAPAHKRGYYGMFTVLGGG
MAV_4627|M.avium_104 AWAPVLLLVLRVLQGFALGGEWGGAVLLAVEHSPGDRRGRYGAVPQVGLA
MLBr_01562|M.leprae_Br4923 IARLLQGIGSAIAS---PTGLALVATTFSKGSARNTATAVFAAMTAIGSV
. * . : . . .
Mflv_1075|M.gilvum_PYR-GCK VGFLLGSVTVTL-------LQALLP--AEAMESYGWRIPFLIAGPLGLVG
Mvan_5748|M.vanbaalenii_PYR-1 LGFLIGSVTVTL-------LQALLP--AAAMESYGWRIPFLIAGPLGLVG
TH_3722|M.thermoresistible__bu LGFLLGSVTVTA-------LQVALP--DQVMTDVGWRIPFLLAGPLGLIG
MAB_2263c|M.abscessus_ATCC_199 GGFAVGAAFVLV-------LDATLS--DDAMHTWGWRIPFLVALPMGLIG
MSMEG_0818|M.smegmatis_MC2_155 LPSVVGGTLILI-------LSSTMP--AAAFESYGWRIPFLLALPLAVFG
Mb1232|M.bovis_AF2122/97 IALVLTSLTFLG-------VNYTIGESSPTFMQWGWRIPFLVSAALIAVA
Rv1200|M.tuberculosis_H37Rv IALVLTSLTFLG-------VNYTIGESSPTFMQWGWRIPFLVSAALIAVA
MMAR_4238|M.marinum_M VALVVSGLTFLG-------VNYTIGESSSAFLQWGWRVPFLLSALLIAFA
MUL_0949|M.ulcerans_Agy99 VALVVSGLTFLG-------VNYTIGESSSALLQWGWRVPFLLSALLIAFA
MAV_4627|M.avium_104 LGLALGTGIFAF-------LQIVLG--PARFLSYGWRIGFLLSVLLVAIG
MLBr_01562|M.leprae_Br4923 MGLVVGGALTEVSWRLAFLVNVPIGLVMIYLARTALHETHKERMKLDAAG
: :. : : . : . : .
Mflv_1075|M.gilvum_PYR-GCK LYIRLRLGDTPQFAELAKAD------KKAESPLREAVA------------
Mvan_5748|M.vanbaalenii_PYR-1 LYIRLRLGDTPQFAELDKAE------KTADSPLREAVT------------
TH_3722|M.thermoresistible__bu LYIRLRLDDTPAFTALSEQE------EVAQSPLREAVQ------------
MAB_2263c|M.abscessus_ATCC_199 WYLRSRLDDPPLFKEMQEQEP--EGRETAFERLRDLVR------------
MSMEG_0818|M.smegmatis_MC2_155 VWIRAKTEESEAFKQVLAKH-----EHKEFSPIRETFR------------
Mb1232|M.bovis_AF2122/97 LYVRFNIDETPVFAR--ERADEKTRLGPAETPIAQVLR------------
Rv1200|M.tuberculosis_H37Rv LYVRFNIDETPVFAR--ERADEKTRLGPAETPIAQVLR------------
MMAR_4238|M.marinum_M LYVRLEINETPVFARAMAQADDQESTAAARAPLAAVLC------------
MUL_0949|M.ulcerans_Agy99 LYVRLEINETPVFARAMAQADDQESTAAARAPLAAVLC------------
MAV_4627|M.avium_104 IVVRLRVEETPAFREL------QDLQAASTVPIRDILRER----------
MLBr_01562|M.leprae_Br4923 AILATLACTAAVFAFSMGPEKGWISLTTIGSGLVALVAAIAFAVVERTAE
: . * : .
Mflv_1075|M.gilvum_PYR-GCK --------TSWRQILQVVGLFIVFNIGYYVVFTFLPTYFIKTLGFTKSYS
Mvan_5748|M.vanbaalenii_PYR-1 --------TSWRQIIQVIGLFIVFNIGYYVVFTFLPTYFIKTLRFSKSEA
TH_3722|M.thermoresistible__bu --------TAWRSILQVIGLMIIFNVAYYVVFAYMPTYLIKELGLSSSDA
MAB_2263c|M.abscessus_ATCC_199 --------DYRRPMVVMMLLVIALNITNYTLLSYQPTYLKTRLGMSETQG
MSMEG_0818|M.smegmatis_MC2_155 --------QNKVRMAQVIFVMGLTAMGFYFLSGYFVSYVQTTGNLSREQS
Mb1232|M.bovis_AF2122/97 --------RQRREIVLAAGSAVCCFGFVYLASTYLASYAQTRLGYSRGSI
Rv1200|M.tuberculosis_H37Rv --------RQRREIVLAAGSAVCCFGFVYLASTYLASYAQTRLGYSRGSI
MMAR_4238|M.marinum_M --------RQRREIVLAAGAMLAAFGCLYLASTFLPSYAQLHLGYSRNLV
MUL_0949|M.ulcerans_Agy99 --------RQRREIVLAAGAMLAAFGCLYLASTFLPSYAQLHLGYSRNLV
MAV_4627|M.avium_104 --------RSRRNTVLGLLSRWAEGSAFNTWGVFAISYATGALGLNRVSV
MLBr_01562|M.leprae_Br4923 NPVVPFDLFRDRNRLVTFIAIFLAGGVIFSLTVSIGLYIQDILGYSALRA
* .
Mflv_1075|M.gilvum_PYR-GCK FLSITLACLVALILILPLAALSDRFGRRPLLIGGAVAFIVLGYPLFLLIT
Mvan_5748|M.vanbaalenii_PYR-1 FVSITLACLVALILILPLAALSDRIGRRPLLIGGAVSFAVLGYPLFLLLT
TH_3722|M.thermoresistible__bu FISSTIACVVALVLILPLAALSDRIGRKPLLVGGAIAFAVLAYPLFLLLN
MAB_2263c|M.abscessus_ATCC_199 QIVILIGEVAMMAFLPFCGALSDRVGRKPMWAFSLTGLLVLSLPMFWLMG
MSMEG_0818|M.smegmatis_MC2_155 LLINAIAMAIYALLLPVGGIIGDRVGRKPMLIAGSAALAVLSIPCFMLVT
Mb1232|M.bovis_AF2122/97 LFDSVLGGLLCIVFTALSSALCDQLGRRRVLLAGWAVALPWSLLVMPLID
Rv1200|M.tuberculosis_H37Rv LFDSVLGGLLCIVFTALSSALCDQLGRRRVLLAGWAVALPWSLLVMPLID
MMAR_4238|M.marinum_M LLIGVLGGLVCIGFVALSAILCDRFGRRRVMLVGWAVGLFWSPLVMPLIG
MUL_0949|M.ulcerans_Agy99 LLIGVLGGLVCIGFVALSAILCDRFGRRRVMLVGWAVGLFWSPLVMPLIG
MAV_4627|M.avium_104 LIAVTIAALLMAVLLPVSGVLTDRFGARRVYLAGIACYGLAAFPAFALFG
MLBr_01562|M.leprae_Br4923 GIGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGWLLMVAMLYGWAFMN
. . : .:.. : : . :.
Mflv_1075|M.gilvum_PYR-GCK SGSPVAAITGHCLLA---------------AIESVYIS-----CAVSAGV
Mvan_5748|M.vanbaalenii_PYR-1 SGSLVAAITAHCLLA---------------AIASVYIS-----SAVSAGV
TH_3722|M.thermoresistible__bu SGSTAAAMIAHAGLA---------------AIESVVIS-----VAVVTAV
MAB_2263c|M.abscessus_ATCC_199 QG-FGWAIVGFAVLG---------------LLYIPQLS-----TITATFP
MSMEG_0818|M.smegmatis_MC2_155 SGSLSLAMVGQLLFV---------------VPLCIYGG-----GCYTFFV
Mb1232|M.bovis_AF2122/97 SGSPSLFAVAVVGMY---------------AIGGFGFG-----PTASFIP
Rv1200|M.tuberculosis_H37Rv SGSPSLFAVAVVGMY---------------AIGGFGFG-----PTASFIP
MMAR_4238|M.marinum_M SGDTVGFTVALLGLY---------------AVAASGFG-----PAASLIP
MUL_0949|M.ulcerans_Agy99 SGDTVGFTVALLGLY---------------AVAASGFG-----PAASLIP
MAV_4627|M.avium_104 TKKLLWFGLAMVIVFG--------------VVHALFYG-----AQGTLYS
MLBr_01562|M.leprae_Br4923 RGVPYFPNLLAPIVVGGIGIGMAVVPLTLSAIAGVGFDRIGPVSAIALML
. .
Mflv_1075|M.gilvum_PYR-GCK ELFATRVRFSGFSVGYNICVAVFGGTT---PYVVTWLTATTG-NSIAPAF
Mvan_5748|M.vanbaalenii_PYR-1 ELFATRIRFSGFSVGYNVCVAVFGGTT---PYVVTWLTAASG-NAIAPAF
TH_3722|M.thermoresistible__bu EQFTTRVRYSGMSVGYNVCVALFGGTT---PYVMTWLIDTTA-NPLAPAC
MAB_2263c|M.abscessus_ATCC_199 CMFPTQVRYAGFAISYNLSTAAFGGTA---PLVNDLVVARTG-WDLFPAA
MSMEG_0818|M.smegmatis_MC2_155 EVFTTATRFTSAAISYNVAYAVFGGTA---PFIGTMLVSSTE-LDAAPGY
Mb1232|M.bovis_AF2122/97 ELFATSYRYTGSALAANLAGVAGGALP---PVIAGALVATYG--SWAIGV
Rv1200|M.tuberculosis_H37Rv ELFATSYRYTGSALAANLAGVAGGALP---PVIAGALVATYG--SWAIGV
MMAR_4238|M.marinum_M ELFATRYRYTGTALALNVAGIVGGAAP---PLIAGTLLASYG--SSAVGL
MUL_0949|M.ulcerans_Agy99 ELFATRYRYTGTALALNVAGIVGGAAP---PLIAGTLLASYG--SSAVGL
MAV_4627|M.avium_104 ALYPTNTRYTGLSFVYQFSGVYASGVT---PMILTALIAALGGAPWLACG
MLBr_01562|M.leprae_Br4923 QSLGGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDAQLRALDHGYTYGLL
: .. . :
Mflv_1075|M.gilvum_PYR-GCK YLIAAAAVSLAAVLTLRETARRPLAQVPYNRADEAPGVALRQHRDR
Mvan_5748|M.vanbaalenii_PYR-1 YLIAAAVVSLAAVLTLRESAGRALAQVQERPANVGSGIHEGEMRR-
TH_3722|M.thermoresistible__bu AVTLAAVVSLGVVMTLRETAARPLAGTP------------------
MAB_2263c|M.abscessus_ATCC_199 YLMVACVVGLCALPFLIETAGASIAGTEIPCNEAAVPTDTVRA---
MSMEG_0818|M.smegmatis_MC2_155 YMATAAAVVLLLLLFARVPETRRRIG--------------------
Mb1232|M.bovis_AF2122/97 MLAILALISLVCTYRLPETAGSALVSR-------------------
Rv1200|M.tuberculosis_H37Rv MLAILALISLVCTYRLPETAGSALVSR-------------------
MMAR_4238|M.marinum_M MMTALVAASLLCTYLLPETAGAPLTAA-------------------
MUL_0949|M.ulcerans_Agy99 MMTALVAASLLCTYLLPETAGAPLTAA-------------------
MAV_4627|M.avium_104 YLVATAVISVLATALIRDRD---LYL--------------------
MLBr_01562|M.leprae_Br4923 WVAGAVVIVGVAALFIGYTPEQVAYAQEVKEAVDAGEL--------
: