For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GLIVPVALLVIWHLVAKRGAFSPSQLPPPGDVLEALGELLRRGDLWTHLQTSVSRVFAGYLAGAAVALVL GSLIGLSATVRRLFAPTVAALRTVPSLAWVPLLLLWFGIGETPKVTLVAIGAFFPIYTTTASALSHVDAQ LLEVGRAYGRRGAALVTTVMLPAAAPALVNGLRLGLANAWLFLVAAELIASSKGLGFMLIDSQNTGRTDI MLLAIGLLAGLGKLTDSLFGIFEARLVRKRS*
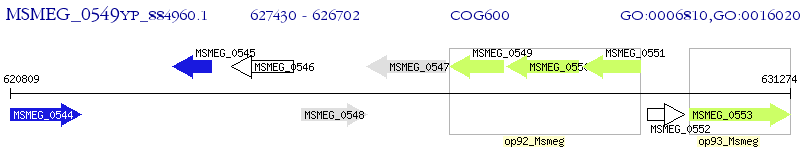
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0549 | - | - | 100% (242) | ABC transporter, permease protein |
| M. smegmatis MC2 155 | MSMEG_3854 | - | 1e-42 | 38.02% (242) | putative aliphatic sulfonates transport permease protein SsuC |
| M. smegmatis MC2 155 | MSMEG_4588 | - | 3e-40 | 38.03% (234) | ABC nitrate/sulfonate/bicarbonate family protein transporter, |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3782c | proZ | 3e-12 | 29.48% (173) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. gilvum PYR-GCK | Mflv_3159 | - | 2e-42 | 37.97% (237) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv3756c | proZ | 3e-12 | 29.48% (173) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2216 | - | 7e-43 | 36.36% (242) | sulfonate ABC transporter, permease protein SsuC |
| M. marinum M | MMAR_5299 | proZ | 2e-13 | 30.64% (173) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. avium 104 | MAV_0143 | - | 3e-95 | 73.71% (232) | putative aliphatic sulfonates transport permease protein SsuC |
| M. thermoresistible (build 8) | TH_4654 | - | 8e-32 | 29.66% (236) | PUTATIVE putative binding-protein-dependent transport systems |
| M. ulcerans Agy99 | MUL_4374 | proZ | 8e-14 | 30.64% (173) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. vanbaalenii PYR-1 | Mvan_3413 | - | 6e-41 | 37.19% (242) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0549|M.smegmatis_MC2_155 ----------------------------------------------MGLI
MAV_0143|M.avium_104 --------------------------------------------------
Mflv_3159|M.gilvum_PYR-GCK MTAVAAPLTTP--RGPAAPAPELVDVIAGHRGR-RSIWG-RIPRPVRRMA
TH_4654|M.thermoresistible__bu VTSLQTARPAPRLRSPAQRRADVTDAQLLHRGEHRTRLGPGRAVPAALAV
MAB_2216|M.abscessus_ATCC_1997 -------MTTR----NTLVAAAATDSDAPESGPSRREWVLERRWEIVRFL
Mvan_3413|M.vanbaalenii_PYR-1 --------MTR----PADPTAPLAQANSGRVG--RR---ADRRWALIRLA
Mb3782c|M.bovis_AF2122/97 --------------------------------------------------
Rv3756c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5299|M.marinum_M --------------------------------------------------
MUL_4374|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_0549|M.smegmatis_MC2_155 VPVALLVIWHLV-AKRGAFSPSQLPPPGDVLEALGELLRRGDLWTHLQTS
MAV_0143|M.avium_104 ------MAWQLATAVTGFFAPSQLPPPGDVVAALSDLARHGELWTHLRAS
Mflv_3159|M.gilvum_PYR-GCK GPVAILAVWTIG-STSGLLNTDLFPPPSDVAVTAWHLLTNGGLALHVGTS
TH_4654|M.thermoresistible__bu GPLLLVALWLIT-SQLGILDPRTLPHPFDIARTAAELWSDGRLPANVAAS
MAB_2216|M.abscessus_ATCC_1997 SPLAVLLIWQAG-SAWGLIDPDILPAPSTIARAGFELIANGQLAEALRVS
Mvan_3413|M.vanbaalenii_PYR-1 SPFILLAVWQLG-SAVGLIPQDVLPAPSLIAEAGIELIGNGQLVDALTVS
Mb3782c|M.bovis_AF2122/97 --------------------MNFLQQALSYLLTASNWTGPVGLAVRTCEH
Rv3756c|M.tuberculosis_H37Rv --------------------MNFLQQALSYLLTASNWTGPVGLAVRTCEH
MMAR_5299|M.marinum_M --------------------MNFLQQALSYLSTAANWTGSAGLAHRTAEH
MUL_4374|M.ulcerans_Agy99 --------------------MNFLQQALSYLSTAANWTGSAGLAHRTAEH
: . : . *
MSMEG_0549|M.smegmatis_MC2_155 VSRVFAGYLAGAAVALVLGSLIGLSATVRRLFAPTVAALRTVPSLAWVPL
MAV_0143|M.avium_104 GWRVLAGYASGAAAGLALGSLVGLSAAARRLLAPTVAAFRTVPSLAWVPL
Mflv_3159|M.gilvum_PYR-GCK ATRVLIGTALGITVGVVLAVLAGLTRTGEDLLDWSMQILKAVPNFALTPL
TH_4654|M.thermoresistible__bu LKLSLISLAIGVAAGLALALLAGLSRIGEALVDGPIQIKRAVPTLAIIPL
MAB_2216|M.abscessus_ATCC_1997 GVRAAEGLLLGGVIGVVLGAAVGLSRLADAVVDPTMQMIRALPHLGLIPL
Mvan_3413|M.vanbaalenii_PYR-1 GLRVVEGLILGGVIGIALGTAVGLSKWFEATVDPPMQMVRALPHLGLIPL
Mb3782c|M.bovis_AF2122/97 LEYTAVAVAASALIAVPVGLLIGHTGRGTLLVVGAVNGLRALPTLGVLLL
Rv3756c|M.tuberculosis_H37Rv LEYTAVAVAASALIAVPVGLLIGHTGRGTLLVVGAVNGLRALPTLGVLLL
MMAR_5299|M.marinum_M LEYTALAVTASALVAIPIGLIIGHTGRGQLLVVSAVNGLRALPTLGVLLL
MUL_4374|M.ulcerans_Agy99 LEYTALAATASALVAIPIGLIIGHTGRGQLLVVSAVNGLRALPTLGVLLL
. . .: :. * : . .: :::* :. *
MSMEG_0549|M.smegmatis_MC2_155 LLLWFGIGETPKVTLVAIGAFFPIYTTTASALSHVDAQLLEVGRAYGRRG
MAV_0143|M.avium_104 LLLWFGIDETPKILMVAVGAFFPVYTTTASALSHIDAHLVEVGRAYGRHG
Mflv_3159|M.gilvum_PYR-GCK LIIWMGIGEGPKIVLITMGVAIAVYINTYSGIRGVDQQLVEMAQTLEARR
TH_4654|M.thermoresistible__bu AIIWFGIGDTMKIVIIATSVLIPVYINTTAQLKGVDLRHVELAQTVGLSR
MAB_2216|M.abscessus_ATCC_1997 FIVWFGIGELPKVLMVALGAAFPLYLNTTSAIRQVDPKLFETAQVLGFSF
Mvan_3413|M.vanbaalenii_PYR-1 FIIWFGIGELPKVLLVALGVSFPLYLNTFSAIRQVDPKLFETAQVLGFSF
Mb3782c|M.bovis_AF2122/97 GVLLFGLGLGPPLVALMLLGIPSLLASTYAGIASVDPLVVDAARAMGMTE
Rv3756c|M.tuberculosis_H37Rv GVLLFGLGLGPPLVALMLLGIPSLLASTYAGIASVDPLVVDAARAMGMTE
MMAR_5299|M.marinum_M GVLLFGLGLGPPIVALMLLGMPPLLAGTYAGIASVDPLVVDAARAMGMTE
MUL_4374|M.ulcerans_Agy99 GVLLFGLGLGPPIVALMLLGMPPLLAGTYAGIASVDPLVVDAARAMGMTE
:: :*:. : : .: * : : :* .: .:.
MSMEG_0549|M.smegmatis_MC2_155 AALVTTVMLPAAAPALVNGLRLGLANAWLFLVAAELIASSKGLGFMLIDS
MAV_0143|M.avium_104 VSLLTAVLLPAAAPELVNGLRLGLANAWLFVVAAELIASSKGLGFLLIDS
Mflv_3159|M.gilvum_PYR-GCK GTLITQVILPGAMPNFLVGLRLGLSSAWLSLIFAEMINTTEGIGFLMSRA
TH_4654|M.thermoresistible__bu VQFIRKVALPGALPGFFTGLRLAVTISWTALVVVEQVNATSGVGFLMTQA
MAB_2216|M.abscessus_ATCC_1997 WQRLRVIVIPGATPQVLVGLRQSLALSWLTLIVAEQINADAGVGFLINNA
Mvan_3413|M.vanbaalenii_PYR-1 WQRFRSIIVPSAAPQVLVGLRQSLAIAWLTLIVAEQINADKGIGFLIMNA
Mb3782c|M.bovis_AF2122/97 SQVLLRVEVPNALPLMLGGLRSATLQVVATATVAAYASLG-GLGGYLIDG
Rv3756c|M.tuberculosis_H37Rv SQVLLRVEVPNALPLMLGGLRSATLQVVATATVAAYASLG-GLGGYLIDG
MMAR_5299|M.marinum_M AQVVLRVEMPNALPLILGGLRSATLQVVATATVAAYASLG-GLGRYLIDG
MUL_4374|M.ulcerans_Agy99 AQVVLRVEMPNALPLILGGLRSATLQVVATVTVAAYASLG-GLGRYLIDG
. : :* * * .. *** . . *:* : .
MSMEG_0549|M.smegmatis_MC2_155 QNTGRTDIMLLAIGLLAGLGKLTDSLFGIFEARLVRKRS-----------
MAV_0143|M.avium_104 QNSGRTDVMLLAIVLLAGLGKLSDAALGAVETRIARRRG-----------
Mflv_3159|M.gilvum_PYR-GCK QTNLQFDVSLLVIVIYAVLGLASYALVRLLERLLLSWRNGFAGFEATA--
TH_4654|M.thermoresistible__bu RLYGQLEVVVVGLVVYAVFGLAGDHLVRTIERRALSWRRALGG-------
MAB_2216|M.abscessus_ATCC_1997 RDFLRIDVIIFGLVVYALLGIVTDAIVRVLERRALRYRS-----------
Mvan_3413|M.vanbaalenii_PYR-1 RDFLRIDIIIFGLIVYALLGIGTDAVVRALERRALRYRS-----------
Mb3782c|M.bovis_AF2122/97 IKERRFHIALVGAMMVAALALTLDGLLALAGWVSVPGTGRMRKL------
Rv3756c|M.tuberculosis_H37Rv IKERRFHIALVGAMMVAALALTLDGLLALAGWVSVPGTGRMRKL------
MMAR_5299|M.marinum_M IKERQFHIALVGALMVAALALSLDGLLALAGWASVPGTGRLSRFWHQSDD
MUL_4374|M.ulcerans_Agy99 IKERQSHIALVGALMVAALALSLDGLLALAGWASVPGTGRVSRFWHQPDD
: .: :. : * :. .
MSMEG_0549|M.smegmatis_MC2_155 -------------------
MAV_0143|M.avium_104 -------------------
Mflv_3159|M.gilvum_PYR-GCK -------------------
TH_4654|M.thermoresistible__bu -------------------
MAB_2216|M.abscessus_ATCC_1997 -------------------
Mvan_3413|M.vanbaalenii_PYR-1 -------------------
Mb3782c|M.bovis_AF2122/97 ---AAVVDKPAAGGGHALR
Rv3756c|M.tuberculosis_H37Rv ---AAVVDKPAAGGGHALR
MMAR_5299|M.marinum_M RIDVAVSPNSAAVGSHVLR
MUL_4374|M.ulcerans_Agy99 RIDVAVSANSAAVGSHVLR