For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTVLPQRAATVPQSPRRRTSTPSPGRRRVILHALQGGIVLSALAAWELGARSGSISSFLFGSPSAVWNVL YKRAVSGELWSDIGVTSSEVILGFLVGAVGGSALGLLLWYSQFVADLTAPFIAAIGSIPVLAVAPLTIIW FGTEMTSKVVIVAFSCVVVSLTTSYSGARRTDPDLVNLMKSFGATRSQIFRKLVVPSAMSWVVSGLKLNI GFALVGAIVGEYISSDAGVGHMILRGSSNFTITLVIAGIAIVMVMVLVFNILVAGLERILGRHQKS*
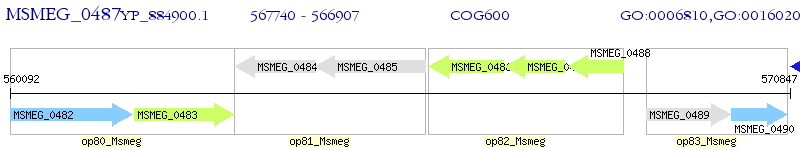
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0487 | - | - | 100% (277) | ABC transporter permease |
| M. smegmatis MC2 155 | MSMEG_2635 | - | 3e-29 | 26.39% (269) | ABC transporter permease protein |
| M. smegmatis MC2 155 | MSMEG_3854 | - | 1e-26 | 28.63% (255) | putative aliphatic sulfonates transport permease protein SsuC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3782c | proZ | 6e-10 | 24.72% (178) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. gilvum PYR-GCK | Mflv_3159 | - | 1e-25 | 28.14% (231) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv3756c | proZ | 6e-10 | 24.72% (178) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2216 | - | 4e-24 | 28.38% (229) | sulfonate ABC transporter, permease protein SsuC |
| M. marinum M | MMAR_5299 | proZ | 1e-08 | 21.28% (188) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. avium 104 | MAV_2433 | - | 7e-26 | 28.09% (235) | putative aliphatic sulfonates transport permease protein SsuC |
| M. thermoresistible (build 8) | TH_4654 | - | 8e-20 | 25.82% (244) | PUTATIVE putative binding-protein-dependent transport systems |
| M. ulcerans Agy99 | MUL_4374 | proZ | 6e-08 | 20.50% (161) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. vanbaalenii PYR-1 | Mvan_3413 | - | 8e-27 | 29.23% (260) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3159|M.gilvum_PYR-GCK MTAVAAPLTTP--RGPAAPAPELVDVIAGHRGR-RSIWG-RIPRPVRRMA
TH_4654|M.thermoresistible__bu VTSLQTARPAPRLRSPAQRRADVTDAQLLHRGEHRTRLGPGRAVPAALAV
MAV_2433|M.avium_104 MSIATALDAAPKRLGGAA-----------SGAPGG--LWARRKWEVVRLV
Mvan_3413|M.vanbaalenii_PYR-1 MTRPADPTAP---LAQAN-----------SGRVG---RRADRRWALIRLA
MAB_2216|M.abscessus_ATCC_1997 MTTRNTLVAAAATDSDAP-----------ESGPSRREWVLERRWEIVRFL
Mb3782c|M.bovis_AF2122/97 --------------------------------------------------
Rv3756c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5299|M.marinum_M --------------------------------------------------
MUL_4374|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_0487|M.smegmatis_MC2_155 MTTVLPQRAATVPQSPRR------------RTSTPSPGRRRVILHALQGG
Mflv_3159|M.gilvum_PYR-GCK GPVAILAVWTIGSTSGLLNTDLFPPPSDVAVTAWHLLTNGGLALHVGTSA
TH_4654|M.thermoresistible__bu GPLLLVALWLITSQLGILDPRTLPHPFDIARTAAELWSDGRLPANVAASL
MAV_2433|M.avium_104 SPLALLALWQLGSAIGVIAQDVLPAPSLILQAGVELTRNGQLADALHIST
Mvan_3413|M.vanbaalenii_PYR-1 SPFILLAVWQLGSAVGLIPQDVLPAPSLIAEAGIELIGNGQLVDALTVSG
MAB_2216|M.abscessus_ATCC_1997 SPLAVLLIWQAGSAWGLIDPDILPAPSTIARAGFELIANGQLAEALRVSG
Mb3782c|M.bovis_AF2122/97 -------------------MNFLQQALSYLLTASNWTGPVGLAVRTCEHL
Rv3756c|M.tuberculosis_H37Rv -------------------MNFLQQALSYLLTASNWTGPVGLAVRTCEHL
MMAR_5299|M.marinum_M -------------------MNFLQQALSYLSTAANWTGSAGLAHRTAEHL
MUL_4374|M.ulcerans_Agy99 -------------------MNFLQQALSYLSTAANWTGSAGLAHRTAEHL
MSMEG_0487|M.smegmatis_MC2_155 IVLSALAAWELGARSGSISSFLFGSPSAVWNVLYKRAVSGELWSDIGVTS
: . . . *
Mflv_3159|M.gilvum_PYR-GCK TRVLIGTALGITVGVVLAVLAGLTRTGEDLLDWSMQILKAVPNFALTPLL
TH_4654|M.thermoresistible__bu KLSLISLAIGVAAGLALALLAGLSRIGEALVDGPIQIKRAVPTLAIIPLA
MAV_2433|M.avium_104 VRVIEGLALGGVIGIVAGAAVGLSRWVEATVDPPLQMIRALPHLGLIPLF
Mvan_3413|M.vanbaalenii_PYR-1 LRVVEGLILGGVIGIALGTAVGLSKWFEATVDPPMQMVRALPHLGLIPLF
MAB_2216|M.abscessus_ATCC_1997 VRAAEGLLLGGVIGVVLGAAVGLSRLADAVVDPTMQMIRALPHLGLIPLF
Mb3782c|M.bovis_AF2122/97 EYTAVAVAASALIAVPVGLLIGHTGRGTLLVVGAVNGLRALPTLGVLLLG
Rv3756c|M.tuberculosis_H37Rv EYTAVAVAASALIAVPVGLLIGHTGRGTLLVVGAVNGLRALPTLGVLLLG
MMAR_5299|M.marinum_M EYTALAVTASALVAIPIGLIIGHTGRGQLLVVSAVNGLRALPTLGVLLLG
MUL_4374|M.ulcerans_Agy99 EYTALAATASALVAIPIGLIIGHTGRGQLLVVSAVNGLRALPTLGVLLLG
MSMEG_0487|M.smegmatis_MC2_155 SEVILGFLVGAVGGSALGLLLWYSQFVADLTAPFIAAIGSIPVLAVAPLT
. . . . : : ::* :.: *
Mflv_3159|M.gilvum_PYR-GCK IIWMGIGEGPKIVLITMGVAIAVYINTYSGIRGVDQQLVEMAQTLEARRG
TH_4654|M.thermoresistible__bu IIWFGIGDTMKIVIIATSVLIPVYINTTAQLKGVDLRHVELAQTVGLSRV
MAV_2433|M.avium_104 ILWFGIGELPKVLLVALGVVFPLYLNTFSAIRQVDPKMLETAQVLGFSFF
Mvan_3413|M.vanbaalenii_PYR-1 IIWFGIGELPKVLLVALGVSFPLYLNTFSAIRQVDPKLFETAQVLGFSFW
MAB_2216|M.abscessus_ATCC_1997 IVWFGIGELPKVLMVALGAAFPLYLNTTSAIRQVDPKLFETAQVLGFSFW
Mb3782c|M.bovis_AF2122/97 VLLFGLGLGPPLVALMLLGIPSLLASTYAGIASVDPLVVDAARAMGMTES
Rv3756c|M.tuberculosis_H37Rv VLLFGLGLGPPLVALMLLGIPSLLASTYAGIASVDPLVVDAARAMGMTES
MMAR_5299|M.marinum_M VLLFGLGLGPPIVALMLLGMPPLLAGTYAGIASVDPLVVDAARAMGMTEA
MUL_4374|M.ulcerans_Agy99 VLLFGLGLGPPIVALMLLGMPPLLAGTYAGIASVDPLVVDAARAMGMTEA
MSMEG_0487|M.smegmatis_MC2_155 IIWFGTEMTSKVVIVAFSCVVVSLTTSYSGARRTDPDLVNLMKSFGATRS
:: :* :: : : : .* .: : .
Mflv_3159|M.gilvum_PYR-GCK TLITQVILPGAMPNFLVGLRLGLSSAWLSLIFAEMINTTEGIGFLMSRAQ
TH_4654|M.thermoresistible__bu QFIRKVALPGALPGFFTGLRLAVTISWTALVVVEQVNATSGVGFLMTQAR
MAV_2433|M.avium_104 QRFRRILVPGTAPQVLVGLRQSLAIAWLTLIVAEQINADKGIGFLINNAR
Mvan_3413|M.vanbaalenii_PYR-1 QRFRSIIVPSAAPQVLVGLRQSLAIAWLTLIVAEQINADKGIGFLIMNAR
MAB_2216|M.abscessus_ATCC_1997 QRLRVIVIPGATPQVLVGLRQSLALSWLTLIVAEQINADAGVGFLINNAR
Mb3782c|M.bovis_AF2122/97 QVLLRVEVPNALPLMLGGLR-SATLQVVATATVAAYASLGGLGGYLIDGI
Rv3756c|M.tuberculosis_H37Rv QVLLRVEVPNALPLMLGGLR-SATLQVVATATVAAYASLGGLGGYLIDGI
MMAR_5299|M.marinum_M QVVLRVEMPNALPLILGGLR-SATLQVVATATVAAYASLGGLGRYLIDGI
MUL_4374|M.ulcerans_Agy99 QVVLRVEMPNALPLILGGLR-SATLQVVATVTVAAYASLGGLGRYLIDGI
MSMEG_0487|M.smegmatis_MC2_155 QIFRKLVVPSAMSWVVSGLKLNIGFALVGAIVGEYISSDAGVGHMILRGS
. : :*.: . .. **: : *:* : .
Mflv_3159|M.gilvum_PYR-GCK TNLQFDVSLLVIVIYAVLGLASYALVRLLERLLLSWRNGFAGFEATA---
TH_4654|M.thermoresistible__bu LYGQLEVVVVGLVVYAVFGLAGDHLVRTIERRALSWRRALGG--------
MAV_2433|M.avium_104 DFLRIDIIIFGLTIYALLGIITDAVVRLIERRAVRYRH------------
Mvan_3413|M.vanbaalenii_PYR-1 DFLRIDIIIFGLIVYALLGIGTDAVVRALERRALRYRS------------
MAB_2216|M.abscessus_ATCC_1997 DFLRIDVIIFGLVVYALLGIVTDAIVRVLERRALRYRS------------
Mb3782c|M.bovis_AF2122/97 KERRFHIALVGAMMVAALALTLDGLLALAGWVSVPGTGRMRKL-------
Rv3756c|M.tuberculosis_H37Rv KERRFHIALVGAMMVAALALTLDGLLALAGWVSVPGTGRMRKL-------
MMAR_5299|M.marinum_M KERQFHIALVGALMVAALALSLDGLLALAGWASVPGTGRLSRFWHQSDDR
MUL_4374|M.ulcerans_Agy99 KERQSHIALVGALMVAALALSLDGLLALAGWASVPGTGRVSRFWHQPDDR
MSMEG_0487|M.smegmatis_MC2_155 SNFTITLVIAGIAIVMVMVLVFNILVAGLERILGRHQKS-----------
: : : : : ::
Mflv_3159|M.gilvum_PYR-GCK ------------------
TH_4654|M.thermoresistible__bu ------------------
MAV_2433|M.avium_104 ------------------
Mvan_3413|M.vanbaalenii_PYR-1 ------------------
MAB_2216|M.abscessus_ATCC_1997 ------------------
Mb3782c|M.bovis_AF2122/97 --AAVVDKPAAGGGHALR
Rv3756c|M.tuberculosis_H37Rv --AAVVDKPAAGGGHALR
MMAR_5299|M.marinum_M IDVAVSPNSAAVGSHVLR
MUL_4374|M.ulcerans_Agy99 IDVAVSANSAAVGSHVLR
MSMEG_0487|M.smegmatis_MC2_155 ------------------