For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VKVVVADDSVLLREGLIRLLTEGGHEVAAAVGDGPALVQAVDTHRPDVSVVDVRMPPSHTDEGLRAAVEA RRRVPRSPILVLSQYVEVSYADDLLADGAGAIGYLLKDRVMALTQFLDAVQRVASGGTVLDPDVVAQLLV RRRRDDPLRQLSDRELQVLGLMAEGRSNTNIARTLVVSDGAVEKHIGNIFAKLALPPDSEQHRRVLAVLA YLRG
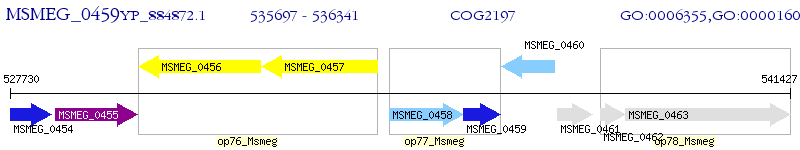
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0459 | - | - | 100% (214) | two-component system response regulator |
| M. smegmatis MC2 155 | MSMEG_4969 | - | 1e-60 | 54.72% (212) | two-component system response regulator |
| M. smegmatis MC2 155 | MSMEG_4378 | - | 1e-54 | 53.30% (212) | two-component system response regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3157c | devR | 7e-21 | 33.17% (205) | two component transcriptional regulatory protein DevR |
| M. gilvum PYR-GCK | Mflv_2280 | - | 2e-59 | 54.25% (212) | two component LuxR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3133c | devR | 7e-21 | 33.17% (205) | two component transcriptional regulatory protein DevR |
| M. leprae Br4923 | MLBr_01286 | - | 5e-11 | 32.71% (107) | putative two-component response regulatory protein |
| M. abscessus ATCC 19977 | MAB_4498c | - | 7e-54 | 50.47% (212) | two-component LuxR family response regulator |
| M. marinum M | MMAR_4782 | narL | 1e-20 | 34.34% (198) | nitrate/nitrite response regulator protein NarL |
| M. avium 104 | MAV_5280 | - | 5e-24 | 36.77% (223) | two-component system response regulator |
| M. thermoresistible (build 8) | TH_1532 | - | 1e-60 | 55.66% (212) | two-component system response regulator |
| M. ulcerans Agy99 | MUL_0351 | narL | 5e-21 | 34.34% (198) | nitrate/nitrite response regulator protein NarL |
| M. vanbaalenii PYR-1 | Mvan_4386 | - | 9e-59 | 56.46% (209) | two component LuxR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2280|M.gilvum_PYR-GCK -------------MRIVIAEDSALLRAGIERILTDAG-HQVVAGVPDATD
Mvan_4386|M.vanbaalenii_PYR-1 ----------------MIAEDSALLRAGIERILTDAG-HQVVAGVPDATN
TH_1532|M.thermoresistible__bu -------------MRIVIAEDSALLRAGIERILTDAG-HEVVAGVPDATD
MSMEG_0459|M.smegmatis_MC2_155 -------------MKVVVADDSVLLREGLIRLLTEGG-HEVAAAVGDGPA
MAB_4498c|M.abscessus_ATCC_199 ----------MSAIRVLVADDDTLLREGVASVLEKAG-FEVVGRAGDATA
Mb3157c|M.bovis_AF2122/97 ------------MVKVFLVDDHEVVRRGLVDLLGADPELDVVGEAGSVAE
Rv3133c|M.tuberculosis_H37Rv ------------MVKVFLVDDHEVVRRGLVDLLGADPELDVVGEAGSVAE
MAV_5280|M.avium_104 ------MTDPAPEITVLLVDDQDLVRSGLRRILRRKDGFVIVAECADGDE
MMAR_4782|M.marinum_M -----MDVTKTEKVRVVVGDDHPLFREGVVRALSLSGSVNVVGEADDGAA
MUL_0351|M.ulcerans_Agy99 -----MDVTKTEKVRVVVGDDHPLFREGVVRALSLSGSVNVVGEADDGAA
MLBr_01286|M.leprae_Br4923 MTGPTTDADAATPHRVLIAEDEALIRLDLAEMLRDEG-YDIVGEAADGQE
.: :* :.* .: * :.. .
Mflv_2280|M.gilvum_PYR-GCK LLRLVNDERPDLAILDVRMPPTFTDEGIRAAALLRSQNPESPVLVLSHYV
Mvan_4386|M.vanbaalenii_PYR-1 LLRLVNDERPDLAILDVRMPPTFTDEGIRAAALLRSQNPESPVLVLSHYV
TH_1532|M.thermoresistible__bu LLRLVNELRPDLAIVDVRMPPTFTDEGIRAAALLRSQNPESPVLVLSHYV
MSMEG_0459|M.smegmatis_MC2_155 LVQAVDTHRPDVSVVDVRMPPSHTDEGLRAAVEARRRVPRSPILVLSQYV
MAB_4498c|M.abscessus_ATCC_199 LLELVRTESPNLVVVDIRMPPTHTTEGLEAAQKIRGEHPGIGVLVLSAHV
Mb3157c|M.bovis_AF2122/97 AMARVPAARPDVAVLDVRLP---DGNGIELCRDLLSRMPDLRCLILTSYT
Rv3133c|M.tuberculosis_H37Rv AMARVPAARPDVAVLDVRLP---DGNGIELCRDLLSRMPDLRCLILTSYT
MAV_5280|M.avium_104 VPAAVAAHRPDVVVMDLRMR---RVDGIEATRRLGGRPP---VLALTTFN
MMAR_4782|M.marinum_M ALELIKTHLPDVALLDYRMP---RMDGAQVAAAVRSNELPTRVLLISAHD
MUL_0351|M.ulcerans_Agy99 ALELIETHLPDVALLDYRMP---RMDGAQVAAAVRSNGLPTRVLLISAHD
MLBr_01286|M.leprae_Br4923 AVELAERHKPDLVIMDVKMP---RRDGIDAASEIASKRIAP-IVVLTAFS
*:: ::* :: :* . : :: .
Mflv_2280|M.gilvum_PYR-GCK EERYAADLIASDTRGFGYLLKDRVADVPAFLDAVEVVGAGGTVLDPEVVS
Mvan_4386|M.vanbaalenii_PYR-1 EERYAADLIASDTRGFGYLLKDRVADVPAFLDAVEVVGSGGTVLDPEVVS
TH_1532|M.thermoresistible__bu EERYAADLIASDTKGFGYLLKDRVADVPAFLDAVEVVGGGGTVLDPEVVS
MSMEG_0459|M.smegmatis_MC2_155 EVSYADDLLADGAGAIGYLLKDRVMALTQFLDAVQRVASGGTVLDPDVVA
MAB_4498c|M.abscessus_ATCC_199 EVEYAAELITSGD-GVGYLLKSRIGDVGEFADACTRVAGGGSIVDPELVR
Mb3157c|M.bovis_AF2122/97 SDEAMLDAILAGA--SGYVVKD--IKGMELARAVKDVGAGRSLLDNRAAA
Rv3133c|M.tuberculosis_H37Rv SDEAMLDAILAGA--SGYVVKD--IKGMELARAVKDVGAGRSLLDNRAAA
MAV_5280|M.avium_104 EDELLSGALRAGA--AGFVLKD--SSAEELIRAVRAVARGEGYLDPAVTS
MMAR_4782|M.marinum_M ESAIVYQALQQGA--AGFLLKD--STRTEIVKAVLDCAKGRDVVAPSLVG
MUL_0351|M.ulcerans_Agy99 ESAIVYQALQQGA--AGFLLKD--STRTEIVKAVLDCAKGRDVVAPSLVG
MLBr_01286|M.leprae_Br4923 QRDLVERARDAGA--MAYLVKP--FTISDLIPAIALAMSRFSELTALERE
. . .:::* : * :
Mflv_2280|M.gilvum_PYR-GCK QILARSHRRNV------LDALTPRENEVLQLMAEGKTNSAIASGLHMSVG
Mvan_4386|M.vanbaalenii_PYR-1 QILVRSRRRNA------LDALTPREHEVLQLMAEGKTNSAIAAALHMSVG
TH_1532|M.thermoresistible__bu QILVRSHRRSA------LDELTAREHDVLRLMAEGKTNSAIAGALHISIG
MSMEG_0459|M.smegmatis_MC2_155 QLLVRRRRDDP------LRQLSDRELQVLGLMAEGRSNTNIARTLVVSDG
MAB_4498c|M.abscessus_ATCC_199 ELLSARRKDDP------LSALSQRELEVLELMAQGLSNSGIAGRLWITES
Mb3157c|M.bovis_AF2122/97 ALMAKLRGAAE--KQDPLSGLTDQERTLLGLLSEGLTNKQIADRMFLAEK
Rv3133c|M.tuberculosis_H37Rv ALMAKLRGAAE--KQDPLSGLTDQERTLLGLLSEGLTNKQIADRMFLAEK
MAV_5280|M.avium_104 RVLTTYRKAAPGPHGAAIAELTTRERDVLTLIGKGLSNSEIADELCISGV
MMAR_4782|M.marinum_M GLAGEIRQRAAP----MAPVLSAREREVLHRIAKGQSIPAIAGELYVAPS
MUL_0351|M.ulcerans_Agy99 GLAGEIRQRAAP----MAPVLSAREREVLHRIAKGQSIPAIAGELYVAPS
MLBr_01286|M.leprae_Br4923 VATLADRLETR-----KLVERAKGLLQVKQGMTEPEAFKWIQRAAMDRRT
: : : : : : *
Mflv_2280|M.gilvum_PYR-GCK SAEKHIASIFTKLGLAPDDTENRRVLAVLRYLES----
Mvan_4386|M.vanbaalenii_PYR-1 SAEKHIASIFTKLDLAPD--ENRRVLAVLRYLDS----
TH_1532|M.thermoresistible__bu SAEKHIASIFTKLGLEPDESENRRVLAVLRYLES----
MSMEG_0459|M.smegmatis_MC2_155 AVEKHIGNIFAKLALPPDSEQHRRVLAVLAYLRG----
MAB_4498c|M.abscessus_ATCC_199 TVEKHVHSVMRRLDLGHADEYHRRVLAVLAFLTHR---
Mb3157c|M.bovis_AF2122/97 TVKNYVSRLLAKLGMERRTQAAVFATELKRSRPPGDGP
Rv3133c|M.tuberculosis_H37Rv TVKNYVSRLLAKLGMERRTQAAVFATELKRSRPPGDGP
MAV_5280|M.avium_104 TVKSHIGRIFGKLDLRDRAAAIVYAYDNGIVAPR----
MMAR_4782|M.marinum_M TVKTHVQRLYEKLGVSDRAAAVAEAMRQGLLD------
MUL_0351|M.ulcerans_Agy99 TVKTHVQRLYEKLGVSDRAAAVAEAMRQGLLD------
MLBr_01286|M.leprae_Br4923 TMKRVAEVVLETLDTPKDT-------------------
: : : *