For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRVDMTPASGEGSRAPEGVNRTVLMVDDDPDVRTSVARGLRHSGFDVRVAASGKEALRLLSTESHDALVL DVQMPELDGVAVVTALRALGNDIPICVLSARDTVNDRIAGLEAGADDYLTKPFDLGELVARLHALLRRAS HTEQPSDTMTVGPVTIDTARRLVFVSGERVDLTKREFDLLAVLAENAGVVLSRQRLLELVWGYDFDVDTN VADVFISYLRRKLERDGVPRVIHTVRGIGYVLREEP
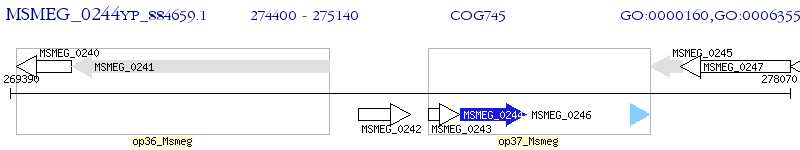
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0244 | - | - | 100% (246) | two component response transcriptional regulatory protein prra |
| M. smegmatis MC2 155 | MSMEG_5662 | prrA | 7e-77 | 65.62% (224) | DNA-binding response regulator PrrA |
| M. smegmatis MC2 155 | MSMEG_5488 | - | 3e-60 | 52.44% (225) | DNA-binding response regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0927c | prrA | 2e-76 | 65.77% (222) | two component response transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_0461 | - | 1e-121 | 89.75% (244) | two component transcriptional regulator |
| M. tuberculosis H37Rv | Rv0903c | prrA | 2e-76 | 65.77% (222) | two component response transcriptional regulatory protein |
| M. leprae Br4923 | MLBr_02123 | - | 3e-75 | 64.86% (222) | two-component response regulator |
| M. abscessus ATCC 19977 | MAB_1926 | - | 9e-99 | 80.53% (226) | putative transcriptional regulatory protein PrrA |
| M. marinum M | MMAR_4633 | prrA | 4e-76 | 65.77% (222) | two-component response transcriptional regulatory protein |
| M. avium 104 | MAV_1022 | - | 3e-76 | 65.77% (222) | two component response transcriptional regulatory protein prra |
| M. thermoresistible (build 8) | TH_4543 | - | 1e-76 | 64.86% (222) | PUTATIVE two component transcriptional regulator, winged helix |
| M. ulcerans Agy99 | MUL_0248 | prrA | 1e-75 | 65.32% (222) | two component response transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_0191 | - | 1e-118 | 88.48% (243) | two component transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4633|M.marinum_M ------------MGGMDTGANSPRVLVVDDDSDVLASLERGLRLSGFEVS
MUL_0248|M.ulcerans_Agy99 ------------MGGMDTGANSPRVLVVDDDSDVLASLERGLRLSGFEVS
MAV_1022|M.avium_104 ---------------MDTAASSPRVLVVDDDSDVLASLERGLRLSGFEVS
MLBr_02123|M.leprae_Br4923 ---------------MDTGVSSPRVLVVDDDSDVLASLERGLRLSGFEVS
Mb0927c|M.bovis_AF2122/97 ------------MGGMDTGVTSPRVLVVDDDSDVLASLERGLRLSGFEVA
Rv0903c|M.tuberculosis_H37Rv ------------MGGMDTGVTSPRVLVVDDDSDVLASLERGLRLSGFEVA
TH_4543|M.thermoresistible__bu --------------------------VVDDDPDVLASLERGLRLSGFEVS
Mflv_0461|M.gilvum_PYR-GCK MRDRVDAPPARGGEPSAHDGAGRTVLMVDDDPDVRTSVARGLRHSGFDVR
Mvan_0191|M.vanbaalenii_PYR-1 ----MDAPSARGGELSAQDGAGRTVLMVDDDPDVRTSVARGLRHSGFDVR
MSMEG_0244|M.smegmatis_MC2_155 --MRVDMTPASGEGSRAPEGVNRTVLMVDDDPDVRTSVARGLRHSGFDVR
MAB_1926|M.abscessus_ATCC_1997 --------------MTQETTARHKVLMVDDDPDVRNSVARGLRHSGFDVR
:****.** *: **** ***:*
MMAR_4633|M.marinum_M TAVDGAEALRSATETRPDAIVLDINMPVLDGVSVVTALRAMDNDVPVCVL
MUL_0248|M.ulcerans_Agy99 TAVDGAEALRSATETRPDAIVLDINMPVLDGVSVVTALRAMDNDVPVCVL
MAV_1022|M.avium_104 TAVDGAEALRSATETRPDAIVLDINMPVLDGVSVVTALRAMDNDVPVCVL
MLBr_02123|M.leprae_Br4923 TAIDGAEALRNATETRPDAIVLDINMPVLDGVSVVTALRAMDNDVPVCVL
Mb0927c|M.bovis_AF2122/97 TAVDGAEALRSATENRPDAIVLDINMPVLDGVSVVTALRAMDNDVPVCVL
Rv0903c|M.tuberculosis_H37Rv TAVDGAEALRSATENRPDAIVLDINMPVLDGVSVVTALRAMDNDVPVCVL
TH_4543|M.thermoresistible__bu TAVDGAEALRSATETRPDAIVLDINMPVLDGVSVVTALRAMDNDVPVCVL
Mflv_0461|M.gilvum_PYR-GCK VAATGKEALRLLSSESHDALVLDVQMPELDGVAVVTALRALGNDIPICVL
Mvan_0191|M.vanbaalenii_PYR-1 VAATGKEALRLLSSESHDALVLDVQMPELDGVAVVTALRALGNDIPICVL
MSMEG_0244|M.smegmatis_MC2_155 VAASGKEALRLLSTESHDALVLDVQMPELDGVAVVTALRALGNDIPICVL
MAB_1926|M.abscessus_ATCC_1997 VAVDGRDALTQLAADEPDALVLDVQMPELDGVAVVTALRALGNDIPICVL
.* * :** : **:***::** ****:*******:.**:*:***
MMAR_4633|M.marinum_M SARSSVDDRVAGLEAGADDYLVKPFVLAELVARVKALLRRRGATATS-SS
MUL_0248|M.ulcerans_Agy99 SARSSVDDRVAGLEAGADDYLVKPFVLAELVARVKALLRRRGATATS-SS
MAV_1022|M.avium_104 SARSSVDDRVAGLEAGADDYLVKPFVLAELVARVKALLRRRGATATS-SS
MLBr_02123|M.leprae_Br4923 SARSSVDDRVAGLEAGADDYLVKPFVLAELVARVKALLRRRGATATS-SS
Mb0927c|M.bovis_AF2122/97 SARSSVDDRVAGLEAGADDYLVKPFVLAELVARVKALLRRRGSTATS-SS
Rv0903c|M.tuberculosis_H37Rv SARSSVDDRVAGLEAGADDYLVKPFVLAELVARVKALLRRRGSTATS-SS
TH_4543|M.thermoresistible__bu SARSSVDDRVAGLEAGADDYLVKPFVLAELVARVKALLRRRGSSATS-SS
Mflv_0461|M.gilvum_PYR-GCK SARDTVNDRIAGLEAGADDYLTKPFDLGELVARLHALLRR--AHHSDPTS
Mvan_0191|M.vanbaalenii_PYR-1 SARDTVNDRIAGLEAGADDYLTKPFDLGELVARLHALLRR--AHHSDPTS
MSMEG_0244|M.smegmatis_MC2_155 SARDTVNDRIAGLEAGADDYLTKPFDLGELVARLHALLRR--ASHTEQPS
MAB_1926|M.abscessus_ATCC_1997 SARDTVNDRIAGLEAGADDYLTKPFDLGELVARLRALLRRRGAGSGGLAD
***.:*:**:***********.*** *.*****::***** : ..
MMAR_4633|M.marinum_M ETITVGPLEVDIPGRRARVNGVDVDLTKREFDLLAVLAEHKTAVLSRAQL
MUL_0248|M.ulcerans_Agy99 EIITVGPLEVDIPGRRARVNGVDVDLTKREFDLLAVLAEHKTAVLSRAQL
MAV_1022|M.avium_104 ETITVGPLEVDIPGRRARVNGVDVDLTKREFDLLAVLAEHKTAVLSRAQL
MLBr_02123|M.leprae_Br4923 ETIAVGPLEVDIPGRRARVNGVDVDLTKREFDLLAVLAEHKTTVLSRAQL
Mb0927c|M.bovis_AF2122/97 ETITVGPLEVDIPGRRARVNGVDVDLTKREFDLLAVLAEHKTAVLSRAQL
Rv0903c|M.tuberculosis_H37Rv ETITVGPLEVDIPGRRARVNGVDVDLTKREFDLLAVLAEHKTAVLSRAQL
TH_4543|M.thermoresistible__bu ETIVVGPLEVDIPGRRARVNGQDVDLTKREFDLLAVLAEHKTAVLSRAQL
Mflv_0461|M.gilvum_PYR-GCK DTMTVGSLTIDTARRLVFVAGERVELTKREFDLLAALAENAGVVLSRQRL
Mvan_0191|M.vanbaalenii_PYR-1 DTMTVGSLTIDTARRLVFVAGERVELTKREFDLLAALAENAGVVLSRQRL
MSMEG_0244|M.smegmatis_MC2_155 DTMTVGPVTIDTARRLVFVSGERVDLTKREFDLLAVLAENAGVVLSRQRL
MAB_1926|M.abscessus_ATCC_1997 SAITVGQITVDVSRRLVFVNGERVELSKREFDLLVVLAENTGVVLSRTRL
. :.** : :* . * . * * *:*:*******..***: .**** :*
MMAR_4633|M.marinum_M LELVWGYDFAADTNVVDVFIGYLRRKLEANGGPRLLHTVRGVGFVLRMQ-
MUL_0248|M.ulcerans_Agy99 LELVWGYDFAADTNVVDVFIGYLRRKLEANGGPRLLHTVRGVGFVLRMH-
MAV_1022|M.avium_104 LELVWGYDFAADTNVVDVFIGYLRRKLEANGGPRLLHTVRGVGFVLRMQ-
MLBr_02123|M.leprae_Br4923 LELVWGYDFAADTNVVDVFIGYLRRKLEANSGPRLLHTVRGVGFVLRMQ-
Mb0927c|M.bovis_AF2122/97 LELVWGYDFAADTNVVDVFIGYLRRKLEAGGGPRLLHTVRGVGFVLRMQ-
Rv0903c|M.tuberculosis_H37Rv LELVWGYDFAADTNVVDVFIGYLRRKLEAGGGPRLLHTVRGVGFVLRMQ-
TH_4543|M.thermoresistible__bu LELVWGYDFAADTNVVDVFIGYLRRKLEATGAPRLLHTVRGVGFVLRTQ-
Mflv_0461|M.gilvum_PYR-GCK LELVWGYDFDVDTNVADVFVSYLRRKLERDGLPRVIHTVRGIGYVLREEA
Mvan_0191|M.vanbaalenii_PYR-1 LELVWGYDFDVDTNVADVFVSYLRRKLEREGLPRVIHTVRGIGYVLRAEL
MSMEG_0244|M.smegmatis_MC2_155 LELVWGYDFDVDTNVADVFISYLRRKLERDGVPRVIHTVRGIGYVLREEP
MAB_1926|M.abscessus_ATCC_1997 LELVWGYDFDVETNVADVFVSYLRRKLEAGGAARVIHTVRGVGYVMREEP
********* .:***.***:.******* . .*::*****:*:*:* .