For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LAKSHLIKPLVVAGAVATAIAAAPVVTADAVAAAGPAAATTAITFLATEQPGGSGCDSNGNCGSGGVNGG PGGGPGGQGCIPGVGCGSGGQFSGPGGVPGGQGCIPGVGCGSGHG*
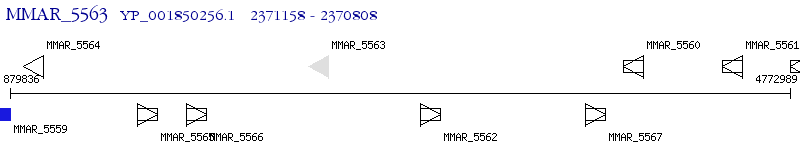
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5563 | - | - | 100% (116) | PE-PGRS family protein |
| M. marinum M | MMAR_2116 | - | 1e-09 | 39.39% (99) | PE-PGRS family protein |
| M. marinum M | MMAR_2823 | - | 2e-09 | 37.86% (103) | PE-PGRS family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3621c | PE_PGRS58 | 2e-08 | 36.26% (91) | PE-PGRS family protein |
| M. gilvum PYR-GCK | Mflv_1054 | - | 2e-13 | 40.48% (126) | hypothetical protein Mflv_1054 |
| M. tuberculosis H37Rv | Rv3514 | PE_PGRS57 | 5e-09 | 47.76% (67) | PE-PGRS family protein |
| M. leprae Br4923 | MLBr_01556 | infB | 1e-06 | 34.12% (85) | translation initiation factor IF-2 |
| M. abscessus ATCC 19977 | MAB_2532 | - | 1e-10 | 43.59% (78) | hypothetical protein MAB_2532 |
| M. avium 104 | MAV_3656 | - | 4e-45 | 74.55% (110) | hypothetical protein MAV_3656 |
| M. smegmatis MC2 155 | MSMEG_5543 | - | 5e-11 | 38.18% (110) | hypothetical protein MSMEG_5543 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4367 | - | 2e-08 | 38.61% (101) | PE-PGRS family protein |
| M. vanbaalenii PYR-1 | Mvan_1264 | - | 5e-08 | 35.54% (121) | triple helix repeat-containing collagen |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5563|M.marinum_M ---------MLAKSHLIKPLVVAGAVATAIAAAPVVTADAVAA-------
MAV_3656|M.avium_104 ----------------MKPLVVAAAAAAAIAAAPAAAADVIVAGP-----
Mb3621c|M.bovis_AF2122/97 -MSFVIVAPEALMSVASEVAGIGSALNAANAAAAAPTTGVLAAAADEVSA
MUL_4367|M.ulcerans_Agy99 -MSFVSVGPGLLEAAARDLAGLGAGLREANLAAAGATVGVLPAAGDEVSA
Rv3514|M.tuberculosis_H37Rv -MSFVLIAPEFVTAAAGDLTNLGSSISAANASAASATTQVLAAGADEVSA
Mvan_1264|M.vanbaalenii_PYR-1 MMFLAPPGGPGPISAAIQLTSTGSEATEPWYVVSFGGSG--APTPNNLAA
Mflv_1054|M.gilvum_PYR-GCK -----------MRYKTSVLGTFGPMLFAGAAATAIATAPLTFAQP-----
MSMEG_5543|M.smegmatis_MC2_155 ---------------------------------------MAMAEP-----
MAB_2532|M.abscessus_ATCC_1997 -------------MHTKARTTVGVAVGSMALIASLIGCGTDAPPRS----
MLBr_01556|M.leprae_Br4923 -MAGKARVHELAKELGVTSKEVLARLNEQGEFVKSASSTVEAPVARRLRE
.
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 AMAALFGAHAQEYQRLSAQAAGFHAQFVQALNAGVNSYASAEAANASPLQ
MUL_4367|M.ulcerans_Agy99 AIAELFGGFGREYQAMGARVALLHDDFVRALTAGGSAYGQAEALNMWPLQ
Rv3514|M.tuberculosis_H37Rv RIAALFGGFGLEYQAISAQVAAYHQRFVQALSTGAGAYASAEAA------
Mvan_1264|M.vanbaalenii_PYR-1 SLVDLSG---TSCGLICNGQDGTGQNGGILFGNGGSGWNSEIAG------
Mflv_1054|M.gilvum_PYR-GCK -----------------APPPCFNADGTPCTALG----------------
MSMEG_5543|M.smegmatis_MC2_155 -----------------APPPCVNADGTPCSDIG----------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 SFGGIKPAADKGAEQVATKAQAKRLGESLDQTLDRALDKAVAGNGATTAA
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 AVEQQVLGLINGPAQTLLGRP---LIGN----------------------
MUL_4367|M.ulcerans_Agy99 ELEQQALGVVNAPSMALFGRP---SIGN----------------------
Rv3514|M.tuberculosis_H37Rv AAEQIVLGVINAPTQALLGRP---LIGDGANATTPGGAGGAGGLLFGNGG
Mvan_1264|M.vanbaalenii_PYR-1 ------VSGGNGGNAGLFGGNGGN--------------------------
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 PVQVDHSAAVVPIVAGEGPSTAHREELAP---------------------
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 -GADGAP-------------------------------------------
MUL_4367|M.ulcerans_Agy99 -GVDGVA-------------------------------------------
Rv3514|M.tuberculosis_H37Rv AGAAGAPGQAGGPGGPAGLWGNGGPGGAGGSGGGTGGAGGAGGWLFGVGG
Mvan_1264|M.vanbaalenii_PYR-1 -GGNGLGS------------------------------------------
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MMAR_5563|M.marinum_M -----------AGPAAATTAITFLA-------------------------
MAV_3656|M.avium_104 -----------AGPAAP-THITFK--------------------------
Mb3621c|M.bovis_AF2122/97 ---------GTGQPGGPGGLL--WGNGGNGGSGVAG--------------
MUL_4367|M.ulcerans_Agy99 ---------GTGQAGGAGGIL--WGNGGNGGLGAAG--------------
Rv3514|M.tuberculosis_H37Rv AGGVGGAGGGTGGAGGPGGLI--WGGGGAGGVGGAGGGTGGAGGRAELLF
Mvan_1264|M.vanbaalenii_PYR-1 ---------GDGGNGGDAGLFSLFGRGGNGGNGGVG--------------
Mflv_1054|M.gilvum_PYR-GCK ----------TAGPGGAAGAIP-GGPAGEAGPG-----------------
MSMEG_5543|M.smegmatis_MC2_155 ----------TAGPGGATGQIP-GGPGGTAGPG-----------------
MAB_2532|M.abscessus_ATCC_1997 ----------VGTTTQTPAPPP----------------------------
MLBr_01556|M.leprae_Br4923 ---------PAGQPSEQPGVPLPGQQGTPAAPHPGHPG------------
.
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 -------------------------------------VGG----------
MUL_4367|M.ulcerans_Agy99 -------------------------------------ANGGAGGSAGLIG
Rv3514|M.tuberculosis_H37Rv GAGGAGGAGTDGGPGATGGTGGHGGVGGDGGWLAPGGAGGAGGQGGAGGA
Mvan_1264|M.vanbaalenii_PYR-1 -------------------------------------IAGATG------A
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MMAR_5563|M.marinum_M ----------------TEQPGGSG-----CDSNGNCGSGGVNG-------
MAV_3656|M.avium_104 -----------------DDPGGGG-----CDANGNCGSGGQDQ-------
Mb3621c|M.bovis_AF2122/97 ---------------PGGSGGAAG----LFGHGGNGGAGGSN--------
MUL_4367|M.ulcerans_Agy99 TGGVGGAGGVGAAGMTGGSGGAGGGGGWLWGAGGSGGAAGAGGSGGSG--
Rv3514|M.tuberculosis_H37Rv GSDGGALGGTGGTGGTGGAGGAGGRGALLLGAGGQGGLGGAGGQGGTGGA
Mvan_1264|M.vanbaalenii_PYR-1 VGAPGVAGATGATGVTSNVTGGNGADAVGYGATGAAGAAGAAGAAG----
Mflv_1054|M.gilvum_PYR-GCK ----------------GASGAIPG------GPAGEAGPGGASG-------
MSMEG_5543|M.smegmatis_MC2_155 ----------------GASGAIPG------GPGGAAGPDGATG-------
MAB_2532|M.abscessus_ATCC_1997 ----------------GGSGSVPG------GPTGGGGPEGGAG-------
MLBr_01556|M.leprae_Br4923 ----------MPTGPHPGPAPKPGGRPPRVGNNPFSSAQSVARPIPRP--
* . . .
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 ------------------------------------------AAGAGGVG
MUL_4367|M.ulcerans_Agy99 -------------------------------GWLWGNAGQTAGAGVGGAG
Rv3514|M.tuberculosis_H37Rv GGDGVLGGVGGTGGKGGVGGVAGLGGAGGAAGQLFSASGAAGNAGVGGAG
Mvan_1264|M.vanbaalenii_PYR-1 -----------------------------------------KAGAVGSAG
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MMAR_5563|M.marinum_M ---------------------------GPGGGPGGQGCIPGVGCGSGGQF
MAV_3656|M.avium_104 ---------------------------GPGGGPGGQGCVPGVGCGSGGQF
Mb3621c|M.bovis_AF2122/97 GAGGAG-------------------WLVGNGGAGGFG--GVGTTVSGNGG
MUL_4367|M.ulcerans_Agy99 GAGGAGDAGDQGVSGVAPGEAGAVGFAGGAGGAGGAGGNGVGGGVNGAGG
Rv3514|M.tuberculosis_H37Rv GQGGDGGAGGAGADADQPGATGGTGFAGGAGGAGGAGGSSGAGGTNGSGG
Mvan_1264|M.vanbaalenii_PYR-1 TAGSAG----------APGVWGNAGLPGAAGAAGGAGAIGTAGVAGAAGT
Mflv_1054|M.gilvum_PYR-GCK --------------------------AIPGGPAGEAGPGGVSGSTNPGGV
MSMEG_5543|M.smegmatis_MC2_155 --------------------------SIPGGPAGTAGPDGATG-VIPGGP
MAB_2532|M.abscessus_ATCC_1997 --------------------------SIPGGPTGGGGPGGGGGEIPGGPT
MLBr_01556|M.leprae_Br4923 -------------PAPRPSASPSSMSPRPGGAVGGGGPRPPRTGVPRPGG
* * *
MMAR_5563|M.marinum_M SGPGG-----------------------VPGGQGCIPGVGCGSGHG----
MAV_3656|M.avium_104 AGPGG-----------------------VPGGQGCLPGVGCGSGHA----
Mb3621c|M.bovis_AF2122/97 AGGAAGAFG-----------------NGGVGGAGGAAVIGGLPGNGGAG-
MUL_4367|M.ulcerans_Agy99 AGGAGGVGGGGGAGAS-GVGAGVAGGDGGTGGAGGAAGVGGAAGTGGTG-
Rv3514|M.tuberculosis_H37Rv AGGQGGAGGAGGAGADNPTGIGGTGGDGGTGGAAGAGGAGGAAGTGGTGG
Mvan_1264|M.vanbaalenii_PYR-1 AGATGGTGVGGNGGAG------QAGGIGGTGAAGAAGQAGTAGETGVTG-
Mflv_1054|M.gilvum_PYR-GCK SGAAG-----------------------PDGVSGCIPNVGCATIPVG---
MSMEG_5543|M.smegmatis_MC2_155 GGTAG-----------------------PGGASGCIPNVGCATIPAP---
MAB_2532|M.abscessus_ATCC_1997 GSGGPG------------------------GGSGCIPNVGCVNVP-----
MLBr_01556|M.leprae_Br4923 GRPGAPVGGRSDAGGG----------NYRGGGVGALPGGGSGGFRGRPGG
. * . *
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 -----GNAGLIGAGGDGGVGGVG---------------------------
MUL_4367|M.ulcerans_Agy99 -----GAAGVAGTGGDGGDGGAGGAGSDASLAGQAG---FAGGSGGAGGV
Rv3514|M.tuberculosis_H37Rv MIGTTGNAGVGGAGGQGGDGGAGGAGADADQPGATGGTGFAGGAGGAGGA
Mvan_1264|M.vanbaalenii_PYR-1 -----GVGGIGGQGTEGGTGGVGGVG----------------------GV
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 GGHGGGGRPGQRGGAAGAFGRPGGAPRRGRKSKRQKRQEYDSMQAPVVGG
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 --APGTNGMNPPPNQ-------------TSQAAN----GSPGANNGAG--
MUL_4367|M.ulcerans_Agy99 GGSAGAGGVNGSGGDGGAGAAGGAGGASATVAGS----GGVGGDGGAGGA
Rv3514|M.tuberculosis_H37Rv GGSSGAGGTNGSGGAGGTGGQGGAGGAGGAGADNPTGIGGTGGDGGTGGA
Mvan_1264|M.vanbaalenii_PYR-1 GATGGAGGVGGKGIQ-------------GGQGLQ----GGQGGDGGTGG-
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 VRLPHGNGETIRLARGASLSDFAEKIDANPAALVQALFNLGEMVTATQSV
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 --------------------------------------SGGAGLPG--NP
MUL_4367|M.ulcerans_Agy99 AGGG-----------------VGAASLGGDGGHAGAGGAGGEGLSGGFNT
Rv3514|M.tuberculosis_H37Rv AGAGGAGGAAGTGGTGGMIGTTGNAGVGGAGGQGGDGGAGGAGADAD-QP
Mvan_1264|M.vanbaalenii_PYR-1 --------------------------------------TGGDATSMG-EA
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 G--------------------------------DETLELLGSEMNYNVQV
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 GAVPGR--AGGAGGLGGSGSDTS-EGPVTG-----GNGGNGGDGGP----
MUL_4367|M.ulcerans_Agy99 GAAGG---IGGAGGAGGTGGDSGGVGGVNGDGGAGGGGGRGGDGGSS---
Rv3514|M.tuberculosis_H37Rv GATGGTGFAGGAGGAGKAGGSSS-AGGTNSSGSAGGTGRQSGTGGAGGAG
Mvan_1264|M.vanbaalenii_PYR-1 GATGGK---GGNGGNGGRGSLFG-RGGNAGNGGNGGTGGNGGNGGTG---
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 VSPEDEDRELLEPFDLTYGEDQGDEDELQVRPPVVTVMGHVDHGKTRLLD
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 ----------GAPGGNGAPGGIG-----VNTG-TG---------------
MUL_4367|M.ulcerans_Agy99 -TSTSGVGGAGGAGGNGGNAGFG-----VSAGGTGGSA--------GDGG
Rv3514|M.tuberculosis_H37Rv ADNPTGIGGTGGDGGTGGAAGAGGAGGAAGTGGTGGMIGTTGNAGVGGAG
Mvan_1264|M.vanbaalenii_PYR-1 -----GTGGNGGTGGTGGTGGIG------GAGSTG---------------
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 TIRKANVREAEAGGITQHIGAYQVGVDLDGSERLITFIDTP---------
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 --------------------------------------------------
MUL_4367|M.ulcerans_Agy99 AGGGAGGVDGVGGASGNGGAAGAGGAGGSGIG-------------AGIDG
Rv3514|M.tuberculosis_H37Rv GSSGAGGTNGSGGAGGTDGQGGAGGAGGAGADNPTGIGGTGGDGGTGGAA
Mvan_1264|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 -----------------------WAYGGNGGNGGDGGAGARG--------
MUL_4367|M.ulcerans_Agy99 GAGGAGGTGGAAGFPGGGGTTGAWGTGGAGGAGGAGGAGDHGGVGVNPGD
Rv3514|M.tuberculosis_H37Rv GAGGAGGAAGTGGTGGMIGTTGNAGVGGAGGQGGDGGAGGAGADADQPGA
Mvan_1264|M.vanbaalenii_PYR-1 ------------------------GTGGTGGTGGTGGTGGTG--------
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 ------------------------GHEAFTAMRARGAKATDIAILVVAAD
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 --------GDGGNGGNG-----LALNGGNGIG------------------
MUL_4367|M.ulcerans_Agy99 SGTAGWAGGAGGAGGAGGSAAFNGVNGAGGAG------------------
Rv3514|M.tuberculosis_H37Rv TGGTGFAGGAGGAGGSGGSSCAGGTNGSGGAGGTCGQVVAGGAGISFSNG
Mvan_1264|M.vanbaalenii_PYR-1 --------GTGGTGGTG------GTGGKGGTG------------------
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 DGVMPQTVEAINHAQAADVPIVVAVNKIDKE-------------------
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 ------------GNGGAGGRG-------GTGAAGGNGGIG--GGATG---
MUL_4367|M.ulcerans_Agy99 ------------GNGGAGGGGGAGASGIGAGVAGGDGGTGGTGGAAG---
Rv3514|M.tuberculosis_H37Rv SNGGTGGTGGVGGTGGDGGNAGTGAGDPGKGGTGGTGGTGGSGGAGGSGG
Mvan_1264|M.vanbaalenii_PYR-1 ------------GTGGDGGTG-------GTGGTGGTGGTGGTGGTGG---
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 ------------GADPAKIRGQLTEYGLVAEDFGGDTMFIDISAKVG---
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 ------------------------------TLTFFGSG------------
MUL_4367|M.ulcerans_Agy99 ------------------------------TGGAAGTGGTGGA--AGAAG
Rv3514|M.tuberculosis_H37Rv ANFNGGTGGTGGTGGKGGLNTDGLSSATSGTGGTGGTGGKGGTGGAGDDS
Mvan_1264|M.vanbaalenii_PYR-1 ------------------------------TGGTGGAG----------DG
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 --GDGGPGGAG--------------ANTAG--------------------
MUL_4367|M.ulcerans_Agy99 SAGDGGAGGAGGAGDDGSAGVVSGDPGTAGKAGG----------------
Rv3514|M.tuberculosis_H37Rv AGGTGGTGGAGGNAGAGGLANTGGTAGNAGIGGDGGQGGNGGQGDSGSGL
Mvan_1264|M.vanbaalenii_PYR-1 LGGTGGTGGTGGFGGQG------GQGGTGG--------------------
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 ------------------------TGGVGGVGGAGGQGGLLFG------D
MUL_4367|M.ulcerans_Agy99 ------AGGAGGVGGNGVAGGVNGSGGAGGAGGAGGTGGAGASGVPASVD
Rv3514|M.tuberculosis_H37Rv GGQPGFAGGAGGKGGAGGSSGAGGTNGSGGAGGAGGQGGAGGAGISFS-N
Mvan_1264|M.vanbaalenii_PYR-1 ------------------------TGGQGGLGGVGGTGGTGGTGG----T
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------TNIEALLEAVLLTADAALDLRANSGMEAQGVAIEAH
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 GGNGGAGGAGGIGGTGASGG----------------AGGKGGSGLVG---
MUL_4367|M.ulcerans_Agy99 GGDGGAGGAGGAAGTGGSGGSAGN----------TGAGGAGGAGGAGGSA
Rv3514|M.tuberculosis_H37Rv GSNGGTGGTGGVGGTGGDGGNAGTGAGDPGKGGTGGTGGTGGSGGAGGSG
Mvan_1264|M.vanbaalenii_PYR-1 GGQGGTGGTGGTGGTGGTGG----------------AGGDGGLGGLGGFG
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 LDRGRGPVATVLVQRGTLRIGDSVVAGDAYGRVRRMVDEHGVDIEAALPS
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 ------GDGGNGGAG----------------GAGGNGGKGGAGGAG----
MUL_4367|M.ulcerans_Agy99 ASTGVNGDGGAGGAGGVGGLGGDSAATAGGGGAGGDGGKGGAGGAGVNGV
Rv3514|M.tuberculosis_H37Rv GANFNGGTGGTGGTGGTGGKGGMGG-IAGDGGPGGDGGNAGVGGKGGTNG
Mvan_1264|M.vanbaalenii_PYR-1 GVGGTGGQGGQGGPGGSTG------------GAGGAGGQAGVAGKG----
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 SPVQVIGFTSVPGAGDNFLVVDEDRIARQIADRRSARKRNALAARSRKRI
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 ----------GGAG-------------MFSQPGVHGAGGTGGQGG-----
MUL_4367|M.ulcerans_Agy99 GGGDGGKGGTGGAGGAGGAG---SDGIIPSNGGLGGNGGNGGNGGQ----
Rv3514|M.tuberculosis_H37Rv NGGSGGTGGTGGAGGNAGAGGLANTGGTAGNAGIGGDGGQGGNGGQGDSG
Mvan_1264|M.vanbaalenii_PYR-1 -------GLFGGSGSAGTAG---VTGASGASGGKGAQGATGAQGATG---
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 SLEDLDSALKETSQLNLILKGDNAGTVEALEEALMGIQVDDEVALRVIDR
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 --------------------------------AGGAGGAGGAAGAGTVVA
MUL_4367|M.ulcerans_Agy99 ------------------------AFGDGVGGAGGIGGKGGNAAIGTIVI
Rv3514|M.tuberculosis_H37Rv SGLGGQPGFAGGPGGKGGAGGNAGTGGTNGSGAGGAGGQGGAGGAGISFS
Mvan_1264|M.vanbaalenii_PYR-1 ----------------------------AQGATGATGASGAQGSAGSAGS
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 G--------------------------VGGITETNVNLASASDAIIIGFN
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 ----GNPGDPGGFGAAGADG--------------LPG-------------
MUL_4367|M.ulcerans_Agy99 NGGVGGAGGSGGLGGATEDG--------------VGGVGCAGGAGGNGGR
Rv3514|M.tuberculosis_H37Rv NGSNGGTGGTGGVGGTGGDGGNAGTGAGDPGKGGTGGTGGTGGSGGAGGS
Mvan_1264|M.vanbaalenii_PYR-1 AGVAGSAGTTGAQGSAGTNG--------------ANGANGSQGAAGKAGA
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 VRAEGKATELASREGVEIRYYLVIYQAIDDIEKALRGMLKPIYEENQ---
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 --------------------------------------------------
MUL_4367|M.ulcerans_Agy99 GDMLFT--------------------------------------------
Rv3514|M.tuberculosis_H37Rv GGANFNGGTGGTGGTGGTGGKGGMGGIAGDGGPGGDGGNAGVGGKGGTNG
Mvan_1264|M.vanbaalenii_PYR-1 TG------------------------------------------------
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 --------------------------------------------------
MUL_4367|M.ulcerans_Agy99 ---VGGSGGTGGDGGGGGSASGSGTGGSGGTGGVGGIGGDGKLGGFGGDG
Rv3514|M.tuberculosis_H37Rv NGGSGGTGGTGGPGGSGGAPTGSGTGGKGGAGGDGGDGADGGAATGVGDG
Mvan_1264|M.vanbaalenii_PYR-1 ---TTGATGAAGDAGDPGSEAVDINGGAGGAGGAGGEGGEGGAGGAGGDG
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 -------LGRAEIRALFRSSKVGIIAGCIISSGVVRRNAKVRLLRDNIVV
MMAR_5563|M.marinum_M --------------------------------------------------
MAV_3656|M.avium_104 --------------------------------------------------
Mb3621c|M.bovis_AF2122/97 --------------------------------------------------
MUL_4367|M.ulcerans_Agy99 GDGGDGGFG--------VIGGQGGGGGFGGLGGTGAGG-APSDKGINGEP
Rv3514|M.tuberculosis_H37Rv GDGGNGGNGGNGGTGVGSPGGLGGAGGTGGLGGAGAGGGADGDDGDDGQP
Mvan_1264|M.vanbaalenii_PYR-1 SEGGAGGSGGAGGSGTGGAGGAGGSGGSGGSGGAGGSGGAAGDSD-----
Mflv_1054|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5543|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 TDNLTVTSLRREKDDVTEVREGFECGMTLGYSDIKEGDVIESYELVEKQR
MMAR_5563|M.marinum_M -----
MAV_3656|M.avium_104 -----
Mb3621c|M.bovis_AF2122/97 -----
MUL_4367|M.ulcerans_Agy99 GNPG-
Rv3514|M.tuberculosis_H37Rv GNNGS
Mvan_1264|M.vanbaalenii_PYR-1 -----
Mflv_1054|M.gilvum_PYR-GCK -----
MSMEG_5543|M.smegmatis_MC2_155 -----
MAB_2532|M.abscessus_ATCC_1997 -----
MLBr_01556|M.leprae_Br4923 A----