For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ASDLRPGPDSPPTDELASAERALAVLQQVLHPIGDGDLFRPTPCPEYDVSGLTGHLLNSIVVLGGMAGAE FTMRDHTSSAEGQVVSAARPALDAWHRRGMEGDVPLGPASMPARVAVSVFSIEFLVHAWDYATAVGHDIH VPDSLADYVLGLAQNLIQPHERGAAGFDDPVQVGDDVSGLHRLVAFTGRNPIR*
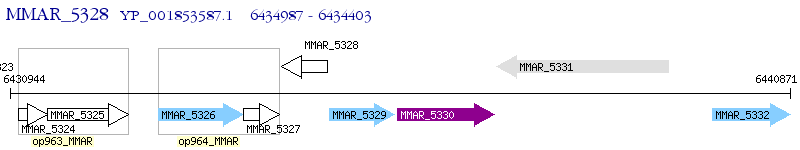
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5328 | - | - | 100% (194) | hypothetical protein MMAR_5328 |
| M. marinum M | MMAR_0942 | - | 6e-41 | 44.50% (191) | transcriptional regulator |
| M. marinum M | MMAR_1076 | - | 2e-09 | 26.11% (180) | hypothetical protein MMAR_1076 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0591 | - | 4e-44 | 47.37% (190) | ArsR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_1118 | - | 1e-08 | 29.31% (174) | hypothetical protein Mflv_1118 |
| M. tuberculosis H37Rv | Rv3773c | - | 1e-69 | 66.49% (194) | hypothetical protein Rv3773c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4446 | - | 2e-32 | 41.48% (176) | hypothetical protein MAB_4446 |
| M. avium 104 | MAV_0248 | - | 1e-64 | 60.91% (197) | hypothetical protein MAV_0248 |
| M. smegmatis MC2 155 | MSMEG_6832 | - | 4e-36 | 40.98% (183) | hypothetical protein MSMEG_6832 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4409 | - | 1e-109 | 97.42% (194) | hypothetical protein MUL_4409 |
| M. vanbaalenii PYR-1 | Mvan_2932 | - | 2e-12 | 28.57% (217) | hypothetical protein Mvan_2932 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5328|M.marinum_M --------------------------------------------------
MUL_4409|M.ulcerans_Agy99 --------------------------------------------------
Rv3773c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0248|M.avium_104 --------------------------------------------------
Mb0591|M.bovis_AF2122/97 MLEVAAEPTRRRLLQLLAPGERTVTQLASQFTVTRSAISQHLGMLAEAGL
MAB_4446|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6832|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2932|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_5328|M.marinum_M --------------------------------------------------
MUL_4409|M.ulcerans_Agy99 --------------------------------------------------
Rv3773c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0248|M.avium_104 --------------------------------------------------
Mb0591|M.bovis_AF2122/97 VTARKQGRERYYRLDERGVLRLRALMESFWSDELDRLVADAAHYPPSQGD
MAB_4446|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6832|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2932|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_5328|M.marinum_M --------------------------------------------------
MUL_4409|M.ulcerans_Agy99 --------------------------------------------------
Rv3773c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0248|M.avium_104 --------------------------------------------------
Mb0591|M.bovis_AF2122/97 CAMPFEKAVVVPLDPTSTFALITQPDRLRRWMAVAARIELRTGGAYRWTV
MAB_4446|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6832|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2932|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_5328|M.marinum_M --------------------------------------------------
MUL_4409|M.ulcerans_Agy99 --------------------------------------------------
Rv3773c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0248|M.avium_104 --------------------------------------------------
Mb0591|M.bovis_AF2122/97 TPGHSAAGTVIDVDPGKRVVFTWGWEDHGDPPPGGSTVTITLTPVDGGTE
MAB_4446|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6832|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2932|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_5328|M.marinum_M -----------------------------MASDLRPGPD-----SPPTDE
MUL_4409|M.ulcerans_Agy99 -----------------------------MASDLRPGPD-----SPPTDE
Rv3773c|M.tuberculosis_H37Rv -----------------------------MPPESRPGPD-----SPPTDE
MAV_0248|M.avium_104 --------------------------MANMAPDLRPGPD-----SPPTDE
Mb0591|M.bovis_AF2122/97 VRLVHDGLTAQQAARHAKGWNHFLDRLVVAGQHGDAGPDEWAAAPDPLDE
MAB_4446|M.abscessus_ATCC_1997 -------------------------------------------MTTALNP
MSMEG_6832|M.smegmatis_MC2_155 ---------------------------MTYAGARASTPH-----WADMDE
Mflv_1118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2932|M.vanbaalenii_PYR-1 -----------------------------MTGLGRHNGAMIAPTSSTTDH
MMAR_5328|M.marinum_M LAS----AERALAVLQQVLHPIGDGDLFRPTPCPEYDVSGLTGHLLNSIV
MUL_4409|M.ulcerans_Agy99 LAS----AERALAVLQQVLHPIGDGDLFRPTPCPEYDVSGLTGHLLNSIV
Rv3773c|M.tuberculosis_H37Rv LAC----AEAALQVLQQVLHTIGRQDKAKQTPCPGYDVKKLTEHLLNSIM
MAV_0248|M.avium_104 LDG----AEAALRVMQQVLHPIAADDMPRPTPCAQFDVTRLTDHLLKSLE
Mb0591|M.bovis_AF2122/97 LSC----AEATLAVLQHVLRGIGASDLTRQTPCTEYDVSQLADHLLRSLA
MAB_4446|M.abscessus_ATCC_1997 LET----VANARAALHEVVSRLTEADNDKQTPNAKFTVAQLTDHLQNSIK
MSMEG_6832|M.smegmatis_MC2_155 LSS----AQSALSALRPVLSAITPDDCGLPTPCAGFDVAALGEHLVGTVR
Mflv_1118|M.gilvum_PYR-GCK MID----MTAACAGTAGLLDRVRDDQLSGATPCSGMDLGALVAHIGGLAL
Mvan_2932|M.vanbaalenii_PYR-1 LKDPRPILDRAIATGGVVIAGVRPEQLTDPTPCTDMDVRALLAHLIGVLA
: : :: : : ** . : * *:
MMAR_5328|M.marinum_M VLGGMAGAEFTMRDH---------TSSAEGQVVSAARPALDAWHR-RGME
MUL_4409|M.ulcerans_Agy99 VLDGMAGAEFTMRDH---------TSTPEGQVVSAARPALDAWHR-RGMD
Rv3773c|M.tuberculosis_H37Rv VLGGMVGAEFSLRAD---------IDSVERLVSGAARSALDAWHR-HGLE
MAV_0248|M.avium_104 ALGGMAGADFPDHADS--------ADSVERRVIAVARPALDAWRQ-RGLD
Mb0591|M.bovis_AF2122/97 IIGAAAGAQLAPRDV---------DAPLETQVADAAQAVMEAWRR-RGLA
MAB_4446|M.abscessus_ATCC_1997 LLGGAAGVDIALTTE----------GSVADRLLPQSQAVVDAWQR-RGID
MSMEG_6832|M.smegmatis_MC2_155 MVGAAADAGFVRFEVVS------HDDDIEARVAASTEAVIAVWQR-RGTA
Mflv_1118|M.gilvum_PYR-GCK AFDAAARKEFGPLTDTPPEMGDGVEDGWRESYPSRLASLASAWREPAAWE
Mvan_2932|M.vanbaalenii_PYR-1 RVAALGNGENPFAVTETS----VADDRWLDAWRQSGRRAADAWSDDAVLE
. . .*
MMAR_5328|M.marinum_M GDVPLGPASMPARVAVSVFSIEFLVHAWDYATAVGHDIHVPDSLADYVLG
MUL_4409|M.ulcerans_Agy99 GDVPLGPASMPARVAVSVFSIEFLVHAWDYATAVGHDIHVPDSLADYVLG
Rv3773c|M.tuberculosis_H37Rv GDVSLGPGSMSAKVAVSVFSVEFLVHAWDYAVAVGSELKAADSLAEYVLE
MAV_0248|M.avium_104 GTVSFGGGEMPARNACAILALELLVHAWDYARAVGRDARAPEPLAEYVLG
Mb0591|M.bovis_AF2122/97 GTVELNSNQVPATVPVGILCLEFLVHAWDFAIATGSQVIASEPVSEYVLA
MAB_4446|M.abscessus_ATCC_1997 GTVTLPIGEYPAEVAVRILGSEFLVHAWDYAVATGQEFEPMDALTDGVLE
MSMEG_6832|M.smegmatis_MC2_155 GEVVFAGRVMPARLALGVLSVELVVHGWDFAQALHRPLGITAAHADFVLA
Mflv_1118|M.gilvum_PYR-GCK GMSRAGGVDFPADVGGLIALTEVVVHGWDVAVSAGLPYEVDGETLAAVHG
Mvan_2932|M.vanbaalenii_PYR-1 RPMALPWIQGSGAEVLASYFSELTVHTWDLATATRQQPDWDDTVVAAAFD
.. *. ** ** * : .
MMAR_5328|M.marinum_M LAQNLIQPH---------------ERGAAGFDDPVQVGDDVSGLHRLVAF
MUL_4409|M.ulcerans_Agy99 LAQNLIQPH---------------ERGAAGFDDPVQVGDDVRGLHRLVAF
Rv3773c|M.tuberculosis_H37Rv LARKLIKPE---------------ERSVAGFNEPVDVPEDGGALERLIAF
MAV_0248|M.avium_104 LAHRVIRPE---------------VRGQAGFDDPVEVPADADALTKLVAF
Mb0591|M.bovis_AF2122/97 VAGKVITPA---------------TRNSAGFAAPAAVGSFAPVLDRLIAF
MAB_4446|M.abscessus_ATCC_1997 SVRMIIQPE---------------RRDGDFFADEVPVPDDSPNLVKLIAF
MSMEG_6832|M.smegmatis_MC2_155 MARHIITPA---------------SRETAGFDDPVSVAADAAALDRLVAY
Mflv_1118|M.gilvum_PYR-GCK HLAAFTADG---------------PVEG-LFDAPVPVPDDAPLLDRVIAM
Mvan_2932|M.vanbaalenii_PYR-1 AARQILPAENRRALYEEISAARGLDEVAVPFAEAVPISDDAPAIDRLVAW
. * . : : :::*
MMAR_5328|M.marinum_M TGRNPIR--------
MUL_4409|M.ulcerans_Agy99 TGRNPIR--------
Rv3773c|M.tuberculosis_H37Rv TGRNPAR--------
MAV_0248|M.avium_104 TGRNPAR--------
Mb0591|M.bovis_AF2122/97 TGRQPTAGHVSAT--
MAB_4446|M.abscessus_ATCC_1997 TGRNPGWSPAS----
MSMEG_6832|M.smegmatis_MC2_155 TGRTPRRWPSNMKSR
Mflv_1118|M.gilvum_PYR-GCK TGRDPSWSAA-----
Mvan_2932|M.vanbaalenii_PYR-1 NGRDPLRRF------
.** *