For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPLENAVVLPVNPAQSFALFTRPEGLRRWMAITARIEPYDAGVYRWTVTPGHTAVGAVVEADPGRRLVLS WGWEGHGDPAPGESTVTVTLAPAPDGTEIRLVHEGLTEQQAARHAEGWSHYLDRLVAAALQGDAGPDEWA AVPDPLDELSCAEATLAVVQHVLRALEPADLARQTPCSQYDVVQLADHLMRSLTIIGSAAGAQLPPRDLD APLETQVADAGQAVLEAWRRRGVDGTVELNANQVPAAVPVGILCLELLVHAWDFAFATDRRVVVSDTVTE YVLTVAGKVITPAARNSAGFAAPVAVGAFAPVLDRLIAFTGRRPVHPTGLTSPIRADPGS
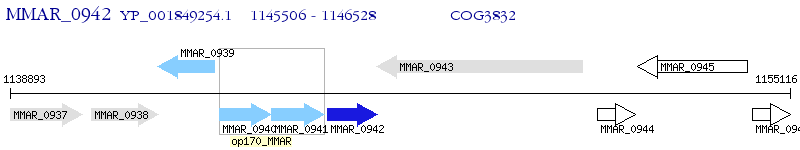
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0942 | - | - | 100% (340) | transcriptional regulator |
| M. marinum M | MMAR_5328 | - | 1e-40 | 44.50% (191) | hypothetical protein MMAR_5328 |
| M. marinum M | MMAR_1076 | - | 2e-07 | 24.44% (180) | hypothetical protein MMAR_1076 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0591 | - | 1e-145 | 74.07% (324) | ArsR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_1118 | - | 1e-06 | 28.42% (183) | hypothetical protein Mflv_1118 |
| M. tuberculosis H37Rv | Rv0576 | - | 1e-145 | 74.07% (324) | ArsR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4446 | - | 1e-29 | 42.16% (185) | hypothetical protein MAB_4446 |
| M. avium 104 | MAV_0248 | - | 2e-40 | 46.07% (191) | hypothetical protein MAV_0248 |
| M. smegmatis MC2 155 | MSMEG_6832 | - | 2e-35 | 39.69% (194) | hypothetical protein MSMEG_6832 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0693 | - | 0.0 | 98.53% (340) | transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_2932 | - | 1e-11 | 29.67% (209) | hypothetical protein Mvan_2932 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0942|M.marinum_M --------------------------------------------------
MUL_0693|M.ulcerans_Agy99 --------------------------MNSPESLDSGYEPAPPGRPAET--
Mb0591|M.bovis_AF2122/97 MLEVAAEPTRRRLLQLLAPGERTVTQLASQFTVTRSAISQHLGMLAEAGL
Rv0576|M.tuberculosis_H37Rv MLEVAAEPTRRRLLQLLAPGERTVTQLASQFTVTRSAISQHLGMLAEAGL
MAV_0248|M.avium_104 --------------------------------------------------
MSMEG_6832|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4446|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_1118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2932|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0942|M.marinum_M --------------------------------------------------
MUL_0693|M.ulcerans_Agy99 ----------------------------------------------VSGE
Mb0591|M.bovis_AF2122/97 VTARKQGRERYYRLDERGVLRLRALMESFWSDELDRLVADAAHYPPSQGD
Rv0576|M.tuberculosis_H37Rv VTARKQGRERYYRLDERGVLRLRALMESFWSDELDRLVADAAHYPPSQGD
MAV_0248|M.avium_104 --------------------------------------------------
MSMEG_6832|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4446|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_1118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2932|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0942|M.marinum_M --MPLENAVVLPVNPAQSFALFTRPEGLRRWMAITARIEPYDAGVYRWTV
MUL_0693|M.ulcerans_Agy99 RAMPLENAVVLPVDPAQSFALFTRPEGLRRWMAITARIEPYDAGVYRWTV
Mb0591|M.bovis_AF2122/97 CAMPFEKAVVVPLDPTSTFALITQPDRLRRWMAVAARIELRTGGAYRWTV
Rv0576|M.tuberculosis_H37Rv CAMPFEKAVVVPLDPTSTFALITQPDRLRRWMAVAARIELRTGGAYRWTV
MAV_0248|M.avium_104 --------------------------------------------------
MSMEG_6832|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4446|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_1118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2932|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0942|M.marinum_M TPGHTAVGAVVEADPGRRLVLSWGWEGHGDPAPGESTVTVTLAPAPDGTE
MUL_0693|M.ulcerans_Agy99 TPGHTAVGAVVEADPGQRLVLSWGWEGHGDPAPGESTVTVTLAPAPDGTE
Mb0591|M.bovis_AF2122/97 TPGHSAAGTVIDVDPGKRVVFTWGWEDHGDPPPGGSTVTITLTPVDGGTE
Rv0576|M.tuberculosis_H37Rv TPGHSAAGTVIDVDPGKRVVFTWGWEDHGDPPPGGSTVTITLTPVDGGTE
MAV_0248|M.avium_104 --------------------------------------------------
MSMEG_6832|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4446|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_1118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2932|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0942|M.marinum_M IRLVHEGLTEQQAARHAEGWSHYLDRLVAAALQGDAGPDEWAAVPDPLDE
MUL_0693|M.ulcerans_Agy99 IRLVHEGLTEQQAARHAEGWSHYLDRLVAAALQGDAGPDEWGAVPGPLDE
Mb0591|M.bovis_AF2122/97 VRLVHDGLTAQQAARHAKGWNHFLDRLVVAGQHGDAGPDEWAAAPDPLDE
Rv0576|M.tuberculosis_H37Rv VRLVHDGLTAQQAARHAKGWNHFLDRLVVAGQRGDAGPDEWAAAPDPLDE
MAV_0248|M.avium_104 --------------------------MANMAPDLRPGPD-----SPPTDE
MSMEG_6832|M.smegmatis_MC2_155 --------------------------MTYAGAR-ASTPH-----WADMDE
MAB_4446|M.abscessus_ATCC_1997 -------------------------------------------MTTALNP
Mflv_1118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2932|M.vanbaalenii_PYR-1 -------------------------MTGLGRHNGAMIAPTSSTTDHLKDP
MMAR_0942|M.marinum_M LSCAEATLAVVQHVLRALEPADLARQTPCSQYDVVQLADHLMRSLTIIGS
MUL_0693|M.ulcerans_Agy99 LSCAEATLAVVQHVLRALEPADLARQTPCSQYDVVQLADHLMRSLTIIGG
Mb0591|M.bovis_AF2122/97 LSCAEATLAVLQHVLRGIGASDLTRQTPCTEYDVSQLADHLLRSLAIIGA
Rv0576|M.tuberculosis_H37Rv LSCAEATLAVLQHVLRGIGASDLTRQTPCTEYDVSQLADHLLRSLAIIGA
MAV_0248|M.avium_104 LDGAEAALRVMQQVLHPIAADDMPRPTPCAQFDVTRLTDHLLKSLEALGG
MSMEG_6832|M.smegmatis_MC2_155 LSSAQSALSALRPVLSAITPDDCGLPTPCAGFDVAALGEHLVGTVRMVGA
MAB_4446|M.abscessus_ATCC_1997 LETVANARAALHEVVSRLTEADNDKQTPNAKFTVAQLTDHLQNSIKLLGG
Mflv_1118|M.gilvum_PYR-GCK MIDMTAACAGTAGLLDRVRDDQLSGATPCSGMDLGALVAHIGGLALAFDA
Mvan_2932|M.vanbaalenii_PYR-1 RPILDRAIATGGVVIAGVRPEQLTDPTPCTDMDVRALLAHLIGVLARVAA
: :: : : ** : : * *: . .
MMAR_0942|M.marinum_M AAGAQLPPR---------DLDAPLETQVADAGQAVLEAWRR-RGVDGTVE
MUL_0693|M.ulcerans_Agy99 AAGAQLPPR---------DLDAPLETQVADAGQAVLEAWRR-RGVDGTVE
Mb0591|M.bovis_AF2122/97 AAGAQLAPR---------DVDAPLETQVADAAQAVMEAWRR-RGLAGTVE
Rv0576|M.tuberculosis_H37Rv AAGAQLAPR---------DVDAPLETQVADAAQAVMEAWRR-RGLAGTVE
MAV_0248|M.avium_104 MAGADFPDHA--------DSADSVERRVIAVARPALDAWRQ-RGLDGTVS
MSMEG_6832|M.smegmatis_MC2_155 AADAGFVRFEVV------SHDDDIEARVAASTEAVIAVWQR-RGTAGEVV
MAB_4446|M.abscessus_ATCC_1997 AAGVDIALT----------TEGSVADRLLPQSQAVVDAWQR-RGIDGTVT
Mflv_1118|M.gilvum_PYR-GCK AARKEFGPLTDTPPEMGDGVEDGWRESYPSRLASLASAWREPAAWEGMSR
Mvan_2932|M.vanbaalenii_PYR-1 LGNGENPFAVTETS----VADDRWLDAWRQSGRRAADAWSDDAVLERPMA
. .*
MMAR_0942|M.marinum_M LNANQVPAAVPVGILCLELLVHAWDFAFATDRRVVVSDTVTEYVLTVAGK
MUL_0693|M.ulcerans_Agy99 LNANQVPAAVPVGILCLELLVHAWDFAFATDRRVVVSDTVTEYVLTVAGK
Mb0591|M.bovis_AF2122/97 LNSNQVPATVPVGILCLEFLVHAWDFAIATGSQVIASEPVSEYVLAVAGK
Rv0576|M.tuberculosis_H37Rv LNSNQVPATVPVGILCLEFLVHAWDFAIATGSQVIASEPVSEYVLAVAGK
MAV_0248|M.avium_104 FGGGEMPARNACAILALELLVHAWDYARAVGRDARAPEPLAEYVLGLAHR
MSMEG_6832|M.smegmatis_MC2_155 FAGRVMPARLALGVLSVELVVHGWDFAQALHRPLGITAAHADFVLAMARH
MAB_4446|M.abscessus_ATCC_1997 LPIGEYPAEVAVRILGSEFLVHAWDYAVATGQEFEPMDALTDGVLESVRM
Mflv_1118|M.gilvum_PYR-GCK AGGVDFPADVGGLIALTEVVVHGWDVAVSAGLPYEVDGETLAAVHGHLAA
Mvan_2932|M.vanbaalenii_PYR-1 LPWIQGSGAEVLASYFSELTVHTWDLATATRQQPDWDDTVVAAAFDAARQ
.. *. ** ** * : .
MMAR_0942|M.marinum_M VITPAA---------------RNSAGFAAPVAVGAFAPVLDRLIAFTGRR
MUL_0693|M.ulcerans_Agy99 VITPAA---------------RNSAGFAAPVAVGAFAPVLDRLIAFTGRR
Mb0591|M.bovis_AF2122/97 VITPAT---------------RNSAGFAAPAAVGSFAPVLDRLIAFTGRQ
Rv0576|M.tuberculosis_H37Rv VITPAT---------------RNSAGFAAPAAVGSFAPVLDRLIAFTGRQ
MAV_0248|M.avium_104 VIRPEV---------------RGQAGFDDPVEVPADADALTKLVAFTGRN
MSMEG_6832|M.smegmatis_MC2_155 IITPAS---------------RETAGFDDPVSVAADAAALDRLVAYTGRT
MAB_4446|M.abscessus_ATCC_1997 IIQPER---------------RDGDFFADEVPVPDDSPNLVKLIAFTGRN
Mflv_1118|M.gilvum_PYR-GCK FTADGP---------------VEG-LFDAPVPVPDDAPLLDRVIAMTGRD
Mvan_2932|M.vanbaalenii_PYR-1 ILPAENRRALYEEISAARGLDEVAVPFAEAVPISDDAPAIDRLVAWNGRD
. * . : : : :::* .**
MMAR_0942|M.marinum_M PVHPTGLTSPIRADPGS
MUL_0693|M.ulcerans_Agy99 PVHPTGLTSPIRADPGS
Mb0591|M.bovis_AF2122/97 PT--AGHVSAT------
Rv0576|M.tuberculosis_H37Rv PT--AGHVSAT------
MAV_0248|M.avium_104 PAR--------------
MSMEG_6832|M.smegmatis_MC2_155 PRRWPSNMKSR------
MAB_4446|M.abscessus_ATCC_1997 PGWSPAS----------
Mflv_1118|M.gilvum_PYR-GCK PSWSAA-----------
Mvan_2932|M.vanbaalenii_PYR-1 PLRRF------------
*