For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTALNPLETVANARAALHEVVSRLTEADNDKQTPNAKFTVAQLTDHLQNSIKLLGGAAGVDIALTTEGS VADRLLPQSQAVVDAWQRRGIDGTVTLPIGEYPAEVAVRILGSEFLVHAWDYAVATGQEFEPMDALTDGV LESVRMIIQPERRDGDFFADEVPVPDDSPNLVKLIAFTGRNPGWSPAS
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
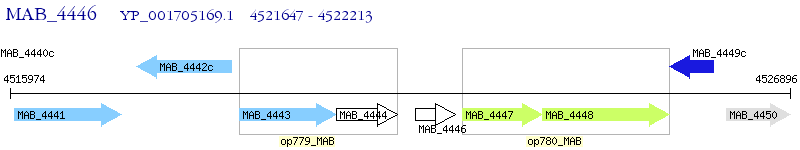
| M. abscessus ATCC 19977 | MAB_4446 | - | - | 100% (188) | hypothetical protein MAB_4446 |
| M. abscessus ATCC 19977 | MAB_2430c | - | 6e-13 | 26.74% (187) | hypothetical protein MAB_2430c |
| M. abscessus ATCC 19977 | MAB_3926 | - | 2e-11 | 29.89% (174) | hypothetical protein MAB_3926 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0591 | - | 3e-29 | 38.80% (183) | ArsR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_1118 | - | 2e-10 | 38.32% (107) | hypothetical protein Mflv_1118 |
| M. tuberculosis H37Rv | Rv3773c | - | 1e-35 | 43.89% (180) | hypothetical protein Rv3773c |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_5328 | - | 3e-32 | 41.48% (176) | hypothetical protein MMAR_5328 |
| M. avium 104 | MAV_0248 | - | 1e-32 | 42.54% (181) | hypothetical protein MAV_0248 |
| M. smegmatis MC2 155 | MSMEG_6832 | - | 4e-23 | 33.51% (188) | hypothetical protein MSMEG_6832 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4409 | - | 1e-31 | 40.91% (176) | hypothetical protein MUL_4409 |
| M. vanbaalenii PYR-1 | Mvan_5694 | - | 3e-11 | 36.57% (134) | hypothetical protein Mvan_5694 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5328|M.marinum_M --------------------------------------------------
MUL_4409|M.ulcerans_Agy99 --------------------------------------------------
Rv3773c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0248|M.avium_104 --------------------------------------------------
Mb0591|M.bovis_AF2122/97 MLEVAAEPTRRRLLQLLAPGERTVTQLASQFTVTRSAISQHLGMLAEAGL
MAB_4446|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6832|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5694|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_5328|M.marinum_M --------------------------------------------------
MUL_4409|M.ulcerans_Agy99 --------------------------------------------------
Rv3773c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0248|M.avium_104 --------------------------------------------------
Mb0591|M.bovis_AF2122/97 VTARKQGRERYYRLDERGVLRLRALMESFWSDELDRLVADAAHYPPSQGD
MAB_4446|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6832|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5694|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_5328|M.marinum_M --------------------------------------------------
MUL_4409|M.ulcerans_Agy99 --------------------------------------------------
Rv3773c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0248|M.avium_104 --------------------------------------------------
Mb0591|M.bovis_AF2122/97 CAMPFEKAVVVPLDPTSTFALITQPDRLRRWMAVAARIELRTGGAYRWTV
MAB_4446|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6832|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5694|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_5328|M.marinum_M --------------------------------------------------
MUL_4409|M.ulcerans_Agy99 --------------------------------------------------
Rv3773c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0248|M.avium_104 --------------------------------------------------
Mb0591|M.bovis_AF2122/97 TPGHSAAGTVIDVDPGKRVVFTWGWEDHGDPPPGGSTVTITLTPVDGGTE
MAB_4446|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6832|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5694|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_5328|M.marinum_M -----------------------------MASDLRPGPD-----SPPTDE
MUL_4409|M.ulcerans_Agy99 -----------------------------MASDLRPGPD-----SPPTDE
Rv3773c|M.tuberculosis_H37Rv -----------------------------MPPESRPGPD-----SPPTDE
MAV_0248|M.avium_104 --------------------------MANMAPDLRPGPD-----SPPTDE
Mb0591|M.bovis_AF2122/97 VRLVHDGLTAQQAARHAKGWNHFLDRLVVAGQHGDAGPDEWAAAPDPLDE
MAB_4446|M.abscessus_ATCC_1997 -------------------------------------------MTTALNP
MSMEG_6832|M.smegmatis_MC2_155 ---------------------------MTYAGARASTPH-----WADMDE
Mflv_1118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5694|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_5328|M.marinum_M LASAERALAVLQQVLHPIGDGDLFRPTPCPEYDVSGLTGHLLNSIVVLGG
MUL_4409|M.ulcerans_Agy99 LASAERALAVLQQVLHPIGDGDLFRPTPCPEYDVSGLTGHLLNSIVVLDG
Rv3773c|M.tuberculosis_H37Rv LACAEAALQVLQQVLHTIGRQDKAKQTPCPGYDVKKLTEHLLNSIMVLGG
MAV_0248|M.avium_104 LDGAEAALRVMQQVLHPIAADDMPRPTPCAQFDVTRLTDHLLKSLEALGG
Mb0591|M.bovis_AF2122/97 LSCAEATLAVLQHVLRGIGASDLTRQTPCTEYDVSQLADHLLRSLAIIGA
MAB_4446|M.abscessus_ATCC_1997 LETVANARAALHEVVSRLTEADNDKQTPNAKFTVAQLTDHLQNSIKLLGG
MSMEG_6832|M.smegmatis_MC2_155 LSSAQSALSALRPVLSAITPDDCGLPTPCAGFDVAALGEHLVGTVRMVGA
Mflv_1118|M.gilvum_PYR-GCK MIDMTAACAGTAGLLDRVRDDQLSGATPCSGMDLGALVAHIGGLALAFDA
Mvan_5694|M.vanbaalenii_PYR-1 MIDMTEACTTTAELLAQVSDDQLTAATPCRDMDLGALIAHVGGLALAFEA
: : :: : : ** : * *: . .
MMAR_5328|M.marinum_M MAGAEFTMRDH---------TSSAEGQVVSAARPALDAWHR-RGMEGDVP
MUL_4409|M.ulcerans_Agy99 MAGAEFTMRDH---------TSTPEGQVVSAARPALDAWHR-RGMDGDVP
Rv3773c|M.tuberculosis_H37Rv MVGAEFSLRAD---------IDSVERLVSGAARSALDAWHR-HGLEGDVS
MAV_0248|M.avium_104 MAGADFPDHADS--------ADSVERRVIAVARPALDAWRQ-RGLDGTVS
Mb0591|M.bovis_AF2122/97 AAGAQLAPRDV---------DAPLETQVADAAQAVMEAWRR-RGLAGTVE
MAB_4446|M.abscessus_ATCC_1997 AAGVDIALTTE----------GSVADRLLPQSQAVVDAWQR-RGIDGTVT
MSMEG_6832|M.smegmatis_MC2_155 AADAGFVRFEVVS------HDDDIEARVAASTEAVIAVWQR-RGTAGEVV
Mflv_1118|M.gilvum_PYR-GCK AARKEFGPLTDTPPEMGDGVEDGWRESYPSRLASLASAWREPAAWEGMSR
Mvan_5694|M.vanbaalenii_PYR-1 AARKDFGPLTDTPPTMEGGVDDGWRQAYPSRLAALAQAWRDPAAWQGMSR
. : . .*: . *
MMAR_5328|M.marinum_M LGPASMPARVAVSVFSIEFLVHAWDYATAVGHDIHVPDSLADYVLGLAQN
MUL_4409|M.ulcerans_Agy99 LGPASMPARVAVSVFSIEFLVHAWDYATAVGHDIHVPDSLADYVLGLAQN
Rv3773c|M.tuberculosis_H37Rv LGPGSMSAKVAVSVFSVEFLVHAWDYAVAVGSELKAADSLAEYVLELARK
MAV_0248|M.avium_104 FGGGEMPARNACAILALELLVHAWDYARAVGRDARAPEPLAEYVLGLAHR
Mb0591|M.bovis_AF2122/97 LNSNQVPATVPVGILCLEFLVHAWDFAIATGSQVIASEPVSEYVLAVAGK
MAB_4446|M.abscessus_ATCC_1997 LPIGEYPAEVAVRILGSEFLVHAWDYAVATGQEFEPMDALTDGVLESVRM
MSMEG_6832|M.smegmatis_MC2_155 FAGRVMPARLALGVLSVELVVHGWDFAQALHRPLGITAAHADFVLAMARH
Mflv_1118|M.gilvum_PYR-GCK AGGVDFPADVGGLIALTEVVVHGWDVAVSAGLPYEVDGETLAAVHGHLAA
Mvan_5694|M.vanbaalenii_PYR-1 AGGVDFPAELGGMIALTEVVVHGWDVAVSAGLRYDVPVDVLNVVLPHVAA
.* : *.:**.** * : *
MMAR_5328|M.marinum_M LIQPHERGAAGFDDPVQVGDDVSGLHRLVAFTGRNPIR--------
MUL_4409|M.ulcerans_Agy99 LIQPHERGAAGFDDPVQVGDDVRGLHRLVAFTGRNPIR--------
Rv3773c|M.tuberculosis_H37Rv LIKPEERSVAGFNEPVDVPEDGGALERLIAFTGRNPAR--------
MAV_0248|M.avium_104 VIRPEVRGQAGFDDPVEVPADADALTKLVAFTGRNPAR--------
Mb0591|M.bovis_AF2122/97 VITPATRNSAGFAAPAAVGSFAPVLDRLIAFTGRQPTAGHVSAT--
MAB_4446|M.abscessus_ATCC_1997 IIQPERRDGDFFADEVPVPDDSPNLVKLIAFTGRNPGWSPAS----
MSMEG_6832|M.smegmatis_MC2_155 IITPASRETAGFDDPVSVAADAAALDRLVAYTGRTPRRWPSNMKSR
Mflv_1118|M.gilvum_PYR-GCK FTADGPVEG-LFDAPVPVPDDAPLLDRVIAMTGRDPSWSAA-----
Mvan_5694|M.vanbaalenii_PYR-1 FAEQGPVEG-LFAAPVPVPGDAPVLHRVLGLTGRDPSWPAG-----
. * . * * :::. *** *