For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LTTLPTPPQTPLVRRPGHQGLVLAVTCLALGTVVAAMASLNVALPDIARQTHADQTQQAWIVDAYSLVFA SLLLPAGAVGDRYGRRLALLVGLVVFGAGSGVATLTTDPTALAGLRALLGVGAALIMPATLSTITSSFPA QRRAAAVSIWTAVAGASGVVGLLASGLLLQWWSWQSIFWLNVLLAAVALTGTLLFVPESAERDPAPLDVV GAVLAAAGIAVLVYAVIEAPAYGWTDPVTVGGIAVGLLILAGFVVIEMGRRYPLLDPRLFTNRRFAAGSL SITLQFFALFGFLFVIMQYLQSVRHYSALTAALGLLAMPIGMIPSSRLSPYLTERFGIRLPWVAGLLIVA EGLMVLAHLDSADPYWHIASGLIPLGAGLGLAMPPATTAITGALPSRLQNVGSAVNDLARELGGALGIAV LGSLLTATYRNHLPTAGAVSAFCTGLHVSLTGAALTIVATAIAVAALLRRPHRPGRHRRSRSAWRGQQRA RAGFTAASDPKRVELFQQRHRDPAGGSQDPARLAHRERLGQCREDLGRPLERVRTEHDRVGDPGQPAGAY RRGQLTRTEPQLTQFVGFRGAKRFGGQPSFEGAHRRVDPRWQARAGRSHQPVGHVQPGSHQQRGNQLGGG *
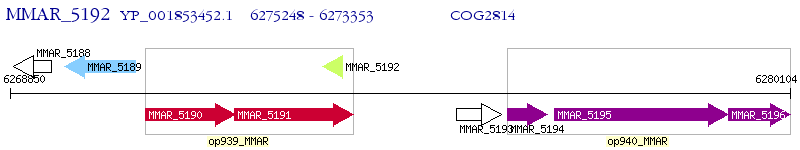
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5192 | - | - | 100% (631) | integral membrane transport protein |
| M. marinum M | MMAR_1146 | - | 4e-94 | 43.32% (434) | integral membrane transport protein |
| M. marinum M | MMAR_5094 | - | 1e-85 | 39.81% (422) | integral membrane transport protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2361c | - | 1e-59 | 34.04% (426) | integral membrane transport protein |
| M. gilvum PYR-GCK | Mflv_5055 | - | 1e-96 | 47.10% (414) | major facilitator transporter |
| M. tuberculosis H37Rv | Rv2333c | - | 1e-58 | 33.80% (426) | integral membrane transport protein |
| M. leprae Br4923 | MLBr_01562 | - | 2e-39 | 29.67% (391) | putative transmembrane efflux protein |
| M. abscessus ATCC 19977 | MAB_0945 | - | 5e-65 | 33.26% (451) | EmrB/QacA family drug resistance transporter |
| M. avium 104 | MAV_1387 | - | 1e-45 | 27.47% (608) | drug transporter |
| M. smegmatis MC2 155 | MSMEG_1427 | - | 1e-101 | 41.86% (485) | transmembrane efflux protein |
| M. thermoresistible (build 8) | TH_2682 | - | 1e-101 | 41.72% (513) | PUTATIVE transmembrane efflux protein |
| M. ulcerans Agy99 | MUL_0911 | - | 1e-94 | 43.05% (439) | integral membrane transport protein |
| M. vanbaalenii PYR-1 | Mvan_0523 | - | 7e-67 | 32.85% (487) | EmrB/QacA family drug resistance transporter |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1427|M.smegmatis_MC2_155 -----------------------------MTDALVEPGQNIDSRMAELSV
TH_2682|M.thermoresistible__bu -----------------------------MVDALSQPGQDIGASVAALST
Mflv_5055|M.gilvum_PYR-GCK -----------------------------MVDTLPRSDQDIDAAVMTLSK
MMAR_5192|M.marinum_M -----------------------------MLTTLPTPPQTP----LVRRP
MUL_0911|M.ulcerans_Agy99 --------------------------------MRIQP----------VFR
Mb2361c|M.bovis_AF2122/97 ------------------------------------------------MN
Rv2333c|M.tuberculosis_H37Rv ------------------------------------------------MN
MAB_0945|M.abscessus_ATCC_1997 ----------------------------------------MEAAVGALSE
Mvan_0523|M.vanbaalenii_PYR-1 ---------------------------------------MNETVVERLSR
MAV_1387|M.avium_104 --------------------------------------------------
MLBr_01562|M.leprae_Br4923 MVTLYDHERAVHNWTAARSDRPAPSRLTQQVEPASERTSKYPTLLPSGRF
MSMEG_1427|M.smegmatis_MC2_155 RQRYWLLIVACLD--VSLVISSMVALNAALPDIATQTSATQAQLTWVIDG
TH_2682|M.thermoresistible__bu RARVWLLTVACMG--VALVISSMVALNAALPDIAVETAATQSQLTWVVDG
Mflv_5055|M.gilvum_PYR-GCK RRRIWLLAVASID--VLMVISSMVALNAALPDIALQTSATQTQLTWIVDG
MMAR_5192|M.marinum_M GHQGLVLAVTCLA--LGTVVAAMASLNVALPDIARQTHADQTQQAWIVDA
MUL_0911|M.ulcerans_Agy99 DHPILALAVICLS--VFVISVDATIVNVALPTLSRELGADTAQLQWIVDA
Mb2361c|M.bovis_AF2122/97 RTQLLTLIATGLG--LFMIFLDALIVNVALPDIQRSFAVGEDGLQWVVAS
Rv2333c|M.tuberculosis_H37Rv RTQLLTLIATGLG--LFMIFLDALIVNVALPDIQRSFAVGEDGLQWVVAS
MAB_0945|M.abscessus_ATCC_1997 RRKIVILGSCCLS--LLIVSMDSTIVNVALPTIRADFGATTSQLQWVIDI
Mvan_0523|M.vanbaalenii_PYR-1 RHKAFVLASCCLS--VLIVSIDVTIANVAIPSIRADLSASPSQMQWVIDI
MAV_1387|M.avium_104 ---------MMIG--FFMIMVDSTIVAIANPTIMADLHIGYDTVVWVTSA
MLBr_01562|M.leprae_Br4923 PSGRFIAAVIVIGGMQLLATMDSTVAIVALPKIQNELSLSDAGRSWGITA
: * * : . *
MSMEG_1427|M.smegmatis_MC2_155 YTLVLACLLLPAGAVGDRYGRRGALLIGLAIFSLASAAPLIFDSPVQLIV
TH_2682|M.thermoresistible__bu YTLVLACLLLPAGAIGDRYGRRGAMLVGLGLFTVASAAPLVLDGPLTLIL
Mflv_5055|M.gilvum_PYR-GCK YTLALACLLLPAGALGDRYGRRGALLVGLAIFALASLAPVFLDSPTQIIV
MMAR_5192|M.marinum_M YSLVFASLLLPAGAVGDRYGRRLALLVGLVVFGAGSGVATLTTDPTALAG
MUL_0911|M.ulcerans_Agy99 YTLVMSGLLLSAGSLSDRYGRRGWLSAGLILFALTSALAAQVNSADTLVA
Mb2361c|M.bovis_AF2122/97 YSLGMAVFIMSAATLADLYGRRRWYLIGVSLFTLGSIACGLAPSIAVLTT
Rv2333c|M.tuberculosis_H37Rv YSLGMAVFIMSAATLADLDGRRRWYLIGVSLFTLGSIACGLAPSIAVLTT
MAB_0945|M.abscessus_ATCC_1997 YTLGLASLLMLSGAAGDRLGRRKVFHLGLSIFAIGSLLCSVAPTVGLLIA
Mvan_0523|M.vanbaalenii_PYR-1 YTLMVASLLLLSGAMADRFGRRRTLQIGLTIFASASLLCSVAPNVETLIG
MAV_1387|M.avium_104 YLLGYAVVLLVAGRLGDRFGTKNLYLIGLAVFTVASVWCGLAGSAAMLIA
MLBr_01562|M.leprae_Br4923 YVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLCAVAWDEVTLVI
* * . .:: .. .* * : *: :* * :
MSMEG_1427|M.smegmatis_MC2_155 ARAVAGAGAAFIMPATLSLLTAAFPKSE-RNKAVGVWAGVVGSGAIVGFL
TH_2682|M.thermoresistible__bu SRMVAGVGAAFVMPATLSLLTAAYPRAQ-RNKAVGIWAAVAGSGAVVGFL
Mflv_5055|M.gilvum_PYR-GCK ARAVAGVGAALIMPATLSLLTSAFPKSE-RNKAVGIWAGVAGSGAVFGFL
MMAR_5192|M.marinum_M LRALLGVGAALIMPATLSTITSSFPAQR-RAAAVSIWTAVAGASGVVGLL
MUL_0911|M.ulcerans_Agy99 ARAAMGVGAAVIFPTTLGLITNIFTDPVPRAKAIGLWTAMVGVGVAVGPI
Mb2361c|M.bovis_AF2122/97 ARGAQGLGAAAVSVTSLALVSAAFPEAKEKARAIGIWTAIASIGTTTGPT
Rv2333c|M.tuberculosis_H37Rv ARGAQGLGAAAVSVTSLALVSAAFPEAKEKARAIGIWTAIASIGTTTGPT
MAB_0945|M.abscessus_ATCC_1997 ARLLQAVGGSMLNPVALSIISQVFTGRVERARALGFWGAVVGISMALGPI
Mvan_0523|M.vanbaalenii_PYR-1 ARFLQAIGGSMLTPAGLSIVTQVFTGRVERARVIGIWGGVVGISMALGPI
MAV_1387|M.avium_104 ARVVQGVGAGVMTPQTLSTITRIFPAER-RGVAVSVWSATAGAASLAGPL
MLBr_01562|M.leprae_Br4923 ARLLQGIGSAIASPTGLALVATTFSKGSARNTATAVFAAMTAIGSVMGLV
* . *.. *. :: :. : . ..: . .. . *
MSMEG_1427|M.smegmatis_MC2_155 VTGGLLHFWTWQSIFVT-FTIASVGLFIVTCTVPSSRDDSAAPLDWLGAA
TH_2682|M.thermoresistible__bu GTGVLLHFWSWPSIFWA-FTVSGAGLFVLACTVASSRDHRATPLDWPGAV
Mflv_5055|M.gilvum_PYR-GCK GTGVLLHFFSWQSIFYM-FAAGALLMFIATCTIGSSRDETATPIDWVGGA
MMAR_5192|M.marinum_M ASGLLLQWWSWQSIFWLNVLLAAVALTGTLLFVPESAERDPAPLDVVGAV
MUL_0911|M.ulcerans_Agy99 TGGWLLEHFSWGSIFLVNVPIAVAAMAGAILFVPTSRDPAAPRVDVPGLM
Mb2361c|M.bovis_AF2122/97 LGGLLVDQWGWRSIFYVNLPMGALVLFLTLCYVEESCNERARRFDLSGQL
Rv2333c|M.tuberculosis_H37Rv LGGLLVDQWGWRSIFYVNLPMGALVLFLTLCYVEESCNERARRFDLSGQL
MAB_0945|M.abscessus_ATCC_1997 VGGLLIQWVGWRAVFWINLPVCVLAFVLTAVFVPETKSATMRDIDPVGQA
Mvan_0523|M.vanbaalenii_PYR-1 MGGILLEYVDWRAVFWINVPICALAVLLTAIFVPESKSVTMRHVDLIGLV
MAV_1387|M.avium_104 AGGVLVDGLGWQWIFFVNVPIGVLGLVLAYWLVP-VLPTQSHRFDLVGVG
MLBr_01562|M.leprae_Br4923 VGGALTEVS-WRLAFLVNVPIGLVMIYLARTALHETHKER-MKLDAAGAI
* * . * * . . : .* *
MSMEG_1427|M.smegmatis_MC2_155 LIGTAVAAFVFGVVEAPVRGWTHPSVWGAMAGGVALAAAFTAVQLRRTHP
TH_2682|M.thermoresistible__bu LIGGAVAVFVYGVLEAPVHGWTHPVVYGSMAAGVALALAFAVVELRSAHP
Mflv_5055|M.gilvum_PYR-GCK LIGTAIAVFVLGVVEAPARGWTDPVVLGCLAAGVVLAVAFAMVQLRREHP
MMAR_5192|M.marinum_M LAAAGIAVLVYAVIEAPAYGWTDPVTVGGIAVGLLILAGFVVIEMGRRYP
MUL_0911|M.ulcerans_Agy99 LSAVGITALVYTIIEAPNWGWTSTRASGGFIAAAIVLVGFALWERRSSHP
Mb2361c|M.bovis_AF2122/97 LFIVAVGALVYAVIEGPQIGWTSVQTIVMLWTAAVGCALFVWLERRSSNP
Rv2333c|M.tuberculosis_H37Rv LFIVAVGALVYAVIEGPQIGWTSVQTIVMLWTAAVGCALFVWLERRSSNP
MAB_0945|M.abscessus_ATCC_1997 LAVLFLFGIVFALIEGPSLGWATPGLIVVLAVALAAFAVFLRYESRRRHP
Mvan_0523|M.vanbaalenii_PYR-1 LGMAFLFGLVFVLIEGPGMGWTDTRVVVSCCLAALAFATFLRYESRRLDP
MAV_1387|M.avium_104 LSGVGMLLIVFGLQQGQAAHWQPWIWALIVAGVGFVTVFVFWQSVNVREP
MLBr_01562|M.leprae_Br4923 LATLACTAAVFAFSMGPEKGWISLTTIGSGLVALVAAIAFAVVERTAENP
* * . . * . . *
MSMEG_1427|M.smegmatis_MC2_155 LLDVRLFRDPNFATGSAGVTFLFFATFGFFFVSMQYIQLIMGYSPIATAI
TH_2682|M.thermoresistible__bu LLDIRLFAKPDFTTGAVGVTFFFLANFGFFFVSMQYMQLMLGYSALQTAF
Mflv_5055|M.gilvum_PYR-GCK LLDVRLFTRPDFATGAVGITFLFMANFGFFFIEMQFMQLVMGYSALETAF
MMAR_5192|M.marinum_M LLDPRLFTNRRFAAGSLSITLQFFALFGFLFVIMQYLQSVRHYSALTAAL
MUL_0911|M.ulcerans_Agy99 MLDVSVFANRRFSGGSLAVTAGFLTLFGFIFVITQYFQFIKDYTALQTGV
Mb2361c|M.bovis_AF2122/97 MMDLTLFRDTSYALAIATICTVFFAVYGMLLLTTQFLQNVRGYTPSVTGL
Rv2333c|M.tuberculosis_H37Rv MMDLTLFRDTSYALAIATICTVFFAVYGMLLLTTQFLQNVRGYTPSVTGL
MAB_0945|M.abscessus_ATCC_1997 FIDLRFFRSIPFASATLTAICACMAWSAFLFTMSLYLQGQRHFSAMHTGL
Mvan_0523|M.vanbaalenii_PYR-1 FVELRFFRSIPFASATAIAVCAFAAWGAFLFMMSIYLQGERGLSAVNTGL
MAV_1387|M.avium_104 LIPLVIFSDRDFSLCNIGVAIISFAATAMMLPLTFYAQAVCGLSPTRSAL
MLBr_01562|M.leprae_Br4923 VVPFDLFRDRNRLVTFIAIFLAGGVIFSLTVSIGLYIQDILGYSALRAGI
.: .* . .: . : * :. :..
MSMEG_1427|M.smegmatis_MC2_155 ALCPLVVPILAFGATTHLYLPLLGLRLTAFLGLLVTAVGLWCMCTLET-G
TH_2682|M.thermoresistible__bu ALTPLTIPLLLLSVTTQWYLPRLGLRAVISTGLLLIAAGLFYARTLEI-A
Mflv_5055|M.gilvum_PYR-GCK ALAPLAFPVLVLGGTLHLYLPKVGLRFAVTGGLLLLAGGLFLMRFLDA-D
MMAR_5192|M.marinum_M GLLAMPIGMIPSSRLSPYLTERFGIRLPWVAGLLIVAEGLMVLAHLDS-A
MUL_0911|M.ulcerans_Agy99 RLLPVAFSIALAAILGPRLVERVGTTAVVAAGLAVFAAALAWASTADA-T
Mb2361c|M.bovis_AF2122/97 MILPFSAAVAIVSPLVGHLVGRIGARVPILAGLCMLMLGLLMLIFSEH-R
Rv2333c|M.tuberculosis_H37Rv MILPFSAAVAIVSPLVGHLVGRIGARVPILAGLCMLMLGLLMLIFSEH-R
MAB_0945|M.abscessus_ATCC_1997 IYLPIAVGALVFSPISGRMVGRFGARPSLLAAGTLFLTASLLLTRVSE-A
Mvan_0523|M.vanbaalenii_PYR-1 MFLPVAVGAFIFSPLSGRLVGRFGARPSLMIAGALIVAATLMFAQMSD-A
MAV_1387|M.avium_104 LIAPMAIANGVFAPFVGKIVDRYHPRPVLGFGFSLLAIALTWLTFEMSPA
MLBr_01562|M.leprae_Br4923 GFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGWLLMVAMLYGWAFMNRG
.. . : .
MSMEG_1427|M.smegmatis_MC2_155 STYFD-FAWPLFIMSIGVGLSTAPVTSAIMGAAPDEKQGVASAVNDTTRE
TH_2682|M.thermoresistible__bu SPYLD-LAVPLLIMSTGIGLCTAPSTSAIMGAVPDEKHGVASAVNDTTRE
Mflv_5055|M.gilvum_PYR-GCK ATFLD-IMWPLLLAASGIGLCTAPTTSAIMNAAPDEKQGVASAVNDATRE
MMAR_5192|M.marinum_M DPYWH-IASGLIPLGAGLGLAMPPATTAITGALPSRLQNVGSAVNDLARE
MUL_0911|M.ulcerans_Agy99 TPYTQ-ITLQMLLLGGGLGLTTAPATEAIMGSLPADRAGVGSAVNDTTRE
Mb2361c|M.bovis_AF2122/97 SSALV-LVG-LGLCGSGVALCLTPITTVAMTAVPAERAGMASGIMSAQRA
Rv2333c|M.tuberculosis_H37Rv SSALV-LVG-LGLCGSGVALCLTPITTVAMTAVPAERAGMASGIMSAQRA
MAB_0945|M.abscessus_ATCC_1997 TPIWE-LLVTFAIFGAGFAMVNAPITNAAVSGMPLDRAGAASAVTSTSRQ
Mvan_0523|M.vanbaalenii_PYR-1 TPQWQ-MLVAFAVFGVGTSMVNAPVTTAAVSGMPTDRAGAAAAITSTSRQ
MAV_1387|M.avium_104 TPIWR-LVLPFFAMGVGMAFVWSPLTATATRNLSAQLAGAGSAVYNSVRQ
MLBr_01562|M.leprae_Br4923 VPYFPNLLAPIVVGGIGIGMAVVPLTLSAIAGVGFDRIGPVSAIALMLQS
. : : . * .: * * . :.: :
MSMEG_1427|M.smegmatis_MC2_155 IGAALGIAVAGSVLAAQYERHLSPA---------LGAMPEAVRGPALNSL
TH_2682|M.thermoresistible__bu IGAALGIAVAGSVLAAEYGDRLRPH---------LGALPEPVRQAASNSL
Mflv_5055|M.gilvum_PYR-GCK IGAALGIAVAGSILAAVYHRSLASR---------LADFPEQIRTTATDSL
MMAR_5192|M.marinum_M LGGALGIAVLGSLLTATYRNHLPTAGA---VSAFCTGLHVSLTGAALTIV
MUL_0911|M.ulcerans_Agy99 LGGTLGVAIAGSVFASVYSGQLGSAS-------ALATLPSDVRSAMKGSM
Mb2361c|M.bovis_AF2122/97 IGSTIGFAVLGSVLAAWLSATLEPH--------LERAVPDPVQRHVLAEI
Rv2333c|M.tuberculosis_H37Rv IGSTIGFAVLGSVLAAWLSATLEPH--------LERAVPDPVQRHVLAEI
MAB_0945|M.abscessus_ATCC_1997 VGVSIGVALCG----------------------SIGTPALWFMCAGLGLL
Mvan_0523|M.vanbaalenii_PYR-1 VGVAIGVAICGPVAGAALGDTNVDF--------AVSAQPLWLVFAGVGVL
MAV_1387|M.avium_104 LGAVLGSAGMAAFMTWRIGAEMPGQPGGGGEDSAGPVLPEFLRGPFAAAM
MLBr_01562|M.leprae_Br4923 LGGPLVLAVIQAVITSRTLYLGGTTG-------PVKFMNDAQLRALDHGY
:* : *
MSMEG_1427|M.smegmatis_MC2_155 AEALTAAEHMG------------------PQGAELA-------QTARQAF
TH_2682|M.thermoresistible__bu AEALEAARQLG------------------PQGARLA-------EFAQSAF
Mflv_5055|M.gilvum_PYR-GCK AHALSISEQMG------------------PQGARLS-------ELAQDAF
MMAR_5192|M.marinum_M ATAIAVAALLRRP--------------HRPGRHRRSRSAWRGQQRARAGF
MUL_0911|M.ulcerans_Agy99 AVAHKVIDQLP------------------AGPAAGVR------DAVNHAF
Mb2361c|M.bovis_AF2122/97 IIDSANPRAHVGG--------------IVPRRHIEHR---DPVAIAEEDF
Rv2333c|M.tuberculosis_H37Rv IIDSANPRAHVGG--------------IVPRRHIEHR---DPVAIAEEDF
MAB_0945|M.abscessus_ATCC_1997 IATLGIVS------------------------------------TSPAAV
Mvan_0523|M.vanbaalenii_PYR-1 IFTLGVYS------------------------------------TSKRAL
MAV_1387|M.avium_104 SQSVLLPAFIALFGIVAALFLVGFRPWAHRDGGTDAFGPDDYGGDYHDDY
MLBr_01562|M.leprae_Br4923 TYGLLWVAG----------------------------------------A
MSMEG_1427|M.smegmatis_MC2_155 VDATGSAVIILG--------------------------------SVVAVA
TH_2682|M.thermoresistible__bu LEAMHVSLIVLG--------------------------------ALCALA
Mflv_5055|M.gilvum_PYR-GCK IHAADRALLVLS--------------------------------LLLLVA
MMAR_5192|M.marinum_M TAASDPKRVELFQQRHRDPAGGSQDPARLAHRERLGQCREDLGRPLERVR
MUL_0911|M.ulcerans_Agy99 LDGLQVATLVCAG--------------------------------IALAA
Mb2361c|M.bovis_AF2122/97 IEGIRVALLVAT----------------------------------ATLA
Rv2333c|M.tuberculosis_H37Rv IEGIRVALLVAT----------------------------------ATLA
MAB_0945|M.abscessus_ATCC_1997 RSAQRLAPLIEG--------------------------------------
Mvan_0523|M.vanbaalenii_PYR-1 RSAEHLAPLIAG--------------------------------------
MAV_1387|M.avium_104 DDDDAYVELILVREPEPEAQQRAGQPRRPQPTPAPPADVRRRDPVESRRS
MLBr_01562|M.leprae_Br4923 VVIVGVAALFIG--------------------------------------
:
MSMEG_1427|M.smegmatis_MC2_155 AVLTGIWGPG------------RDGQQLRVIRRLR--SR-----------
TH_2682|M.thermoresistible__bu AAFVAVWAPG------------RDGRQYGFVERWT--SRWSGEEAGGAVA
Mflv_5055|M.gilvum_PYR-GCK AVFVAIWSPG------------RDGRQWSVVRRLR--KN-----------
MMAR_5192|M.marinum_M TEHDRVGDPGQPAGAYRRGQLTRTEPQLTQFVGFRGAKRFGGQPSFEGAH
MUL_0911|M.ulcerans_Agy99 AIVVGWLLPA------------RAGQPTPAQTEPALAQV-----------
Mb2361c|M.bovis_AF2122/97 VVFLAGWRWFP-----------RDVQTAGSDAKR--EAAYSDDRRVRG--
Rv2333c|M.tuberculosis_H37Rv VVFLAGWRWFP-----------RDVHTAGSDLSERLPTAMTVECAVSHMP
MAB_0945|M.abscessus_ATCC_1997 ----------------------TTAR-EMSRVG-----------------
Mvan_0523|M.vanbaalenii_PYR-1 ----------------------TDPRREAADVA-----------------
MAV_1387|M.avium_104 VLDERPAQVQPIGFAHNGSHVDGGKRLRQVAVRRAPKPGPPADRFTRPPR
MLBr_01562|M.leprae_Br4923 ----------------------YTPEQVAYAQEVKEAVDAGEL-------
MSMEG_1427|M.smegmatis_MC2_155 ----------------------------------------
TH_2682|M.thermoresistible__bu EHDDGGVRAATGDGWEHGTVDDP-----------------
Mflv_5055|M.gilvum_PYR-GCK ----------------------------------------
MMAR_5192|M.marinum_M RRVDPRWQARAGRSHQPVGHVQPGSHQQRGNQLGGG----
MUL_0911|M.ulcerans_Agy99 ----------------------------------------
Mb2361c|M.bovis_AF2122/97 ----------------------------------------
Rv2333c|M.tuberculosis_H37Rv GATWCRLWPA------------------------------
MAB_0945|M.abscessus_ATCC_1997 ----------------------------------------
Mvan_0523|M.vanbaalenii_PYR-1 ----------------------------------------
MAV_1387|M.avium_104 RHHPGPAGHHLGEGESLRGQHHRPDPDDDPTGYGRHSSGN
MLBr_01562|M.leprae_Br4923 ----------------------------------------