For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AGGTQGVAAIVPAAGSGQRLAAGVPKAFYQLEGQTLVERAVRGLLESGVVDTVVVAVPPDRTDEAKLILG REATIVAGGADRTESVSRALSALSAGVSPEFVLVHDAARALTPPALVVRLVDMLRAGHAAVVPVLPLTDT IKAVDANGVVLGTPERAGLRAVQTPQGFSTELLLRAYRHAGVAAFTDDASLVEHVGGQVHVVAGDPLAFK ITNRLDLLLAHAVVRG*
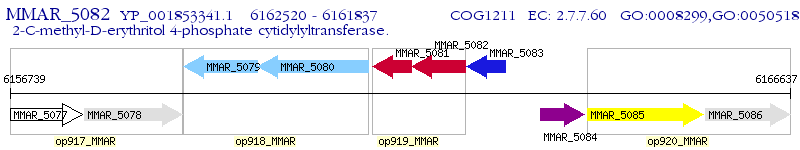
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5082 | ispD | - | 100% (227) | 4-diphosphocytidyl-2C-methyl-D-erythritol synthase IspD |
| M. marinum M | MMAR_4467 | glmU | 1e-05 | 26.34% (186) | UDP-N-acetylglucosamine pyrophosphorylase GlmU |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3613c | ispD | 1e-91 | 76.34% (224) | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
| M. gilvum PYR-GCK | Mflv_2528 | - | 5e-17 | 32.08% (212) | 4-diphosphocytidyl-2C-methyl-D-erythritol synthase |
| M. tuberculosis H37Rv | Rv3582c | ispD | 1e-91 | 76.34% (224) | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
| M. leprae Br4923 | MLBr_00321 | ispD | 2e-81 | 67.92% (240) | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
| M. abscessus ATCC 19977 | MAB_0569 | - | 4e-57 | 54.37% (206) | putative 2-C-methyl-D-erythritol 4-phosphate |
| M. avium 104 | MAV_0571 | ispD | 7e-84 | 71.56% (225) | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
| M. smegmatis MC2 155 | MSMEG_6076 | ispD | 1e-69 | 64.52% (217) | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
| M. thermoresistible (build 8) | TH_2770 | - | 1e-26 | 39.81% (211) | PUTATIVE putative 4-diphosphocytidyl-2C-methyl-D-erythritol |
| M. ulcerans Agy99 | MUL_4158 | ispD | 1e-121 | 98.68% (227) | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
| M. vanbaalenii PYR-1 | Mvan_4130 | - | 4e-17 | 32.30% (226) | 4-diphosphocytidyl-2C-methyl-D-erythritol synthase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5082|M.marinum_M MAGGTQ---GVAAIVPAAGSGQRLAAGVPKAFYQLEGQTLVERAVRGLLE
MUL_4158|M.ulcerans_Agy99 MAGGTQ---GVAAIVPAAGSGQRLAAGVPKAFYQLEGQTLVERAVRGLFE
Mb3613c|M.bovis_AF2122/97 MVREAG---EVVAIVPAAGSGERLAVGVPKAFYQLDGQTLIERAVDGLLD
Rv3582c|M.tuberculosis_H37Rv MVREAG---EVVAIVPAAGSGERLAVGVPKAFYQLDGQTLIERAVDGLLD
MLBr_00321|M.leprae_Br4923 MAVATG---TVVAVVPAAGAGKRLAAGIPKAFCELDGRTLVERAVVGLLE
MAV_0571|M.avium_104 MAPETGSSGAVAAVVPAAGSGERLAAGIPKAFCEIDGASMLARAVAGLLD
MSMEG_6076|M.smegmatis_MC2_155 MA--------TVAVVPAAGSGERLRAGRPKAFVTLGGTPLLEHALSGLRA
MAB_0569|M.abscessus_ATCC_1997 ---------------------MRLGANTPKAFVSVGGRTMLQRCVDGLLA
Mflv_2528|M.gilvum_PYR-GCK -----------MTITALLPVPADLARQPETVFAPVAGQSSLERIVRSLAA
Mvan_4130|M.vanbaalenii_PYR-1 -----------MTVTAILPVPESYVQRREAVFAPVAGESPLVRIVRALAA
TH_2770|M.thermoresistible__bu -----------MTYAAIVPLAATVPR--TDVFTPLAGMPALVRAVRMLAE
.* : * . : : : *
MMAR_5082|M.marinum_M SGVVDTVVVAVPPDRTDEAKLILGR--------EATIVAGGADRTESVSR
MUL_4158|M.ulcerans_Agy99 SGVVDTVVVAVPPDRTDEAKLILGR--------EATIVAGGADRTESVSR
Mb3613c|M.bovis_AF2122/97 SGVVDTVVVAVPADRTDEARQILGH--------RAMIVAGGSNRTDTVNL
Rv3582c|M.tuberculosis_H37Rv SGVVDTVVVAVPADRTDEARQILGH--------RAMIVAGGSNRTDTVNL
MLBr_00321|M.leprae_Br4923 SGVVDHVVVAVPADRIAQTQWVLSQRLANSAGQHATVVAGGADRTKSVCQ
MAV_0571|M.avium_104 SKVVDHVVVAVPADRVDEAKRLLAA--------QATVVAGGADRTASVRL
MSMEG_6076|M.smegmatis_MC2_155 SGVIDRIVIAVPPALTDESKLVFGG-------EDSVIVSGGVDRTESVAL
MAB_0569|M.abscessus_ATCC_1997 SGAVDEVVVVVGADQLERAAALVGS--------GATVVQGGAERTDSVRA
Mflv_2528|M.gilvum_PYR-GCK VC---DVVVAVAEEMSDDVAAVLSAP--DVQAVRLTVAAAPGRRRECLAA
Mvan_4130|M.vanbaalenii_PYR-1 VG---DVVVAAAQSLADEVRETLAAQ--ALPAVRTVGARSPGARADCIAA
TH_2770|M.thermoresistible__bu EAGADVVLVAVAAALADEARAALSG----VAGTEVVSVDGAGRRLDCLHA
:::.. .. . . * :
MMAR_5082|M.marinum_M ALSALSAG---VSPEFVLVHDAARALTPPALVVRLVDMLRAGHAAVVPVL
MUL_4158|M.ulcerans_Agy99 ALSALSAG---VSPEFVLVHDAARALTPPALVVRLVDMLRAGHAAVVPVL
Mb3613c|M.bovis_AF2122/97 ALAVLSGT---AEPEFVLVHDAARALTPPALVARVVEALRDGYAAVVPVL
Rv3582c|M.tuberculosis_H37Rv ALTVLSGT---AEPEFVLVHDAARALTPPALVARVVEALRDGYAAVVPVL
MLBr_00321|M.leprae_Br4923 ALATLPAPSRVGAPEFILVHDAARALTPARLIVRVVDALRAGHTAVVPAL
MAV_0571|M.avium_104 ALAAVP-----GNPAFVLVHDAARALTPPALIARVVQALRDGHRAVVPAL
MSMEG_6076|M.smegmatis_MC2_155 ALEAAG------DAEFVLVHDAARALTPPALIARVVAALKEGHSAVVPGL
MAB_0569|M.abscessus_ATCC_1997 GLAAVD------AADWILVHDAARPLTPPDMIARIVAELRTGRGAVIPAV
Mflv_2528|M.gilvum_PYR-GCK GLAGLHDG-------PVLVHDIGWPIFSQQTLEGVLGALHRGADAVLPIC
Mvan_4130|M.vanbaalenii_PYR-1 GLAALGDGD--G---PVLLHDLTWPLIADATLDAVLSAVHGGADAVLPIR
TH_2770|M.thermoresistible__bu GLDAVRNRS--TPPHHVLVHDCRRPLTPPAVVRRVRAALADGAEAVIPAL
.* :*:** .: . : : : * **:*
MMAR_5082|M.marinum_M PLTDTIKAVDANGVVLGTPERAGLRAVQTPQGFSTELLLRAYRHAG----
MUL_4158|M.ulcerans_Agy99 PLTDTIKAVDANGVVLGTPERAGLRAVQTPQGFSTELLLRAYRHAG----
Mb3613c|M.bovis_AF2122/97 PLSDTIKAVDANGVVLGTPERAGLRAVQTPQGFTTDLLLRSYQRGSLDLP
Rv3582c|M.tuberculosis_H37Rv PLSDTIKAVDANGVVLGTPERAGLRAVQTPQGFTTDLLLRSYQRGSLDLP
MLBr_00321|M.leprae_Br4923 PLSDTIKAVDANGMVLGTPARVGLRAVQTPQGFATELLWCAYQRGP-HLD
MAV_0571|M.avium_104 PLHDTVKAVDANGVVLGTPERDGLRAVQTPQGFATDLLLRAYAAG---AG
MSMEG_6076|M.smegmatis_MC2_155 APADTIKAVDANGAVLGTPERAGLRAVQTPQGFHADVLRRAYARAT----
MAB_0569|M.abscessus_ATCC_1997 AVADTIKSIDVHGDVIGTPDRAGLRAVQTPQGFAADVLRRAYAAAG----
Mflv_2528|M.gilvum_PYR-GCK AVTDSIKAVDGRGVITATVDRAPLRTVQYPRGFSAAMLAQLIGTSD----
Mvan_4130|M.vanbaalenii_PYR-1 PVTDSVKAVDARGAVTATLDRSPLRTVQFPRGFDAAVLERLLADGA----
TH_2770|M.thermoresistible__bu PFTDSAKTVDASGSITATLDRATLRTAQHPRGFPVDRLAQLLDGAD----
. *: *::* * : .* * **:.* *:** . * .
MMAR_5082|M.marinum_M VAAFTDDASLVEHVGGQVHVVAGDPLAFKITNRLDLLLAHAVVRG-----
MUL_4158|M.ulcerans_Agy99 VSAFTDDASLVEHVGGQVHVVGGDPLAFKITNRLDLLLAHAVVRG-----
Mb3613c|M.bovis_AF2122/97 AAEYTDDASLVEHIGGQVQVVDGDPLAFKITTKLDLLLAQAIVRG-----
Rv3582c|M.tuberculosis_H37Rv AAEYTDDASLVEHIGGQVQVVDGDPLAFKITTKLDLLLAQAIVRG-----
MLBr_00321|M.leprae_Br4923 AVDFTDDASLVEHLGGQVQVVAGDPLAFKITTQLDLLLAKKILRR-----
MAV_0571|M.avium_104 TAGFTDDASLVEHVGGQVQVVDGDPLAFKITTQLDLLLAETIVRR-----
MSMEG_6076|M.smegmatis_MC2_155 AGGVTDDASLVEQLGTPVQIVDGDPLAFKITTPLDLVLAEAVLARGA---
MAB_0569|M.abscessus_ATCC_1997 -DIATDDAALVEQIGERVHVVEGDRLAFKITTTLDMTLAEAILGATEGVQ
Mflv_2528|M.gilvum_PYR-GCK --FDDDELETVLTGGDGTVLIDGDTAISRIELPCDAAYLDALLTSRDSDP
Mvan_4130|M.vanbaalenii_PYR-1 --ADFDELGAALSVGTPLTLVEGDTDTMSFDLPADSAFLAAVIAGR----
TH_2770|M.thermoresistible__bu --SAADELLEAQRVGLTLTLVDGDPDGFVVRLPDAASLVEAVITARAGRI
*: . * :: ** . ::
MMAR_5082|M.marinum_M --
MUL_4158|M.ulcerans_Agy99 --
Mb3613c|M.bovis_AF2122/97 --
Rv3582c|M.tuberculosis_H37Rv --
MLBr_00321|M.leprae_Br4923 --
MAV_0571|M.avium_104 --
MSMEG_6076|M.smegmatis_MC2_155 --
MAB_0569|M.abscessus_ATCC_1997 --
Mflv_2528|M.gilvum_PYR-GCK AG
Mvan_4130|M.vanbaalenii_PYR-1 --
TH_2770|M.thermoresistible__bu --