For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTFRGDTAVVVLAAGAGTRMRSDIPKVLHTLAGRSMLSHSLHAMAKLAPQHLVVVLGHCHERISPLVDEL AESLGRTINVTLQDRPLGTGDAVRCGLSALPADYSGIVVVTAGDTPLLDAETLADLIETHSATSSAVTVL TTTLSDPHGYGRILRTQDNAVMAIVEQTDATPSQREIREVNAGVYAFEITALQSALSRLSSDNAQQELYL PDVIAILRRDGQTVSARHIDDSALVAGVNDRVQLAQLGAELNRRIVAAHQMAGVTVIDPATTWIDVDVAI GRDTVIQPGTQLLGHTQIGDRCEIGPDTTLTDVTVGDNASVVRTHGSSSSIGAAAAVGPFTFLRPGTVLG TGGKLGAFVETKNSTIGAGTKVPHLTYVGDADIGDDSNIGAGSVFVNYDGMTKNRATIGSHVRSGAGTRF VAPVNVGDGAYTGAGTVIRDDVPPGALAVSGGPQRNIEDWVQQKRPGTPSAEAARKASAEQSTPPPDADQ TP
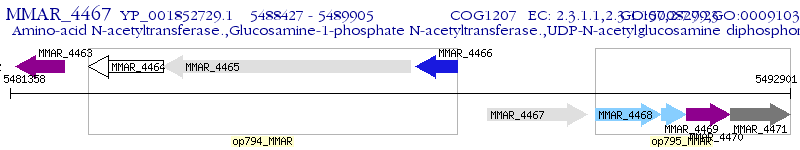
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4467 | glmU | - | 100% (492) | UDP-N-acetylglucosamine pyrophosphorylase GlmU |
| M. marinum M | MMAR_1277 | manB | 3e-19 | 25.75% (369) | d-alpha-D-mannose-1-phosphate guanylyltransferase ManB |
| M. marinum M | MMAR_3646 | cysE | 4e-06 | 31.87% (91) | serine acetyltransferase CysE |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1046c | glmU | 0.0 | 81.01% (495) | UDP-N-acetylglucosamine pyrophosphorylase glmU |
| M. gilvum PYR-GCK | Mflv_1944 | glmU | 0.0 | 72.61% (482) | UDP-N-acetylglucosamine pyrophosphorylase |
| M. tuberculosis H37Rv | Rv1018c | glmU | 0.0 | 80.81% (495) | UDP-N-acetylglucosamine pyrophosphorylase glmU |
| M. leprae Br4923 | MLBr_00249 | glmU | 0.0 | 79.71% (478) | bifunctional N-acetylglucosamine-1-phosphate |
| M. abscessus ATCC 19977 | MAB_1148c | glmU | 0.0 | 70.13% (472) | UDP-N-acetylglucosamine pyrophosphorylase (GlmU) |
| M. avium 104 | MAV_1157 | glmU | 0.0 | 78.41% (477) | UDP-N-acetylglucosamine pyrophosphorylase |
| M. smegmatis MC2 155 | MSMEG_5426 | glmU | 0.0 | 71.16% (482) | UDP-N-acetylglucosamine pyrophosphorylase |
| M. thermoresistible (build 8) | TH_0911 | glmU | 0.0 | 71.28% (484) | Probable UDP-N-acetylglucosamine pyrophosphorylase glmU |
| M. ulcerans Agy99 | MUL_4637 | glmU | 0.0 | 98.78% (492) | UDP-N-acetylglucosamine pyrophosphorylase GlmU |
| M. vanbaalenii PYR-1 | Mvan_4782 | glmU | 0.0 | 73.00% (474) | UDP-N-acetylglucosamine pyrophosphorylase |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_1148c|M.abscessus_ATCC_199 MPGNTAGTAVIVLAAGAGTRMCSDIPKVLHTLGGRSMLAHAVYAAASVNP
MSMEG_5426|M.smegmatis_MC2_155 MTASTE-AAVVVLAAGAGTRMRSDTPKVLHTLAGRGMLAHVLHTVSEIDA
Mflv_1944|M.gilvum_PYR-GCK ----MSDAAVVILAAGAGTRMKSDTPKVLHSLAGRSMLAHSMHAVAGLGP
Mvan_4782|M.vanbaalenii_PYR-1 ----MRDAAVVILAAGAGTRMKSDTPKVLHTLAGRSMLSHALHAVAGLEA
TH_0911|M.thermoresistible__bu ---------VVVLAAGAGTRMRSDTPKVLHTLGGRSMLAHVLHSVARIDP
MMAR_4467|M.marinum_M -MTFRGDTAVVVLAAGAGTRMRSDIPKVLHTLAGRSMLSHSLHAMAKLAP
MUL_4637|M.ulcerans_Agy99 -MTFRGDTAVVVLAAGAGTRMRSDTPKVLHTLAGRSMLSHSLHAMAKLAP
Mb1046c|M.bovis_AF2122/97 -MTFPGDTAVLVLAAGPGTRMRSDTPKVLHTLAGRSMLSHVLHAIAKLAP
Rv1018c|M.tuberculosis_H37Rv -MTFPGDTAVLVLAAGPGTRMRSDTPKVLHTLAGRSMLSHVLHAIAKLAP
MAV_1157|M.avium_104 -MSSPGDTAVLVLAAGPGTRMRSDTPKVLHTLGGRSMLSHLLHAIAKVAP
MLBr_00249|M.leprae_Br4923 -MTFRGDTAVLVLAAGPGSRMRSDTPKVLHTIAGRSMLSHSLHAITKLAP
*::****.*:** ** *****::.**.**:* ::: : : .
MAB_1148c|M.abscessus_ATCC_199 EHLVIVLGHERERIATAVDILAESLGRRIEIAVQEQQLGTGHAVACGLQA
MSMEG_5426|M.smegmatis_MC2_155 RHLVAVLGHDRERIAPEVARLSEELGRAIDVAVQDQQLGTGHAVNCGLTA
Mflv_1944|M.gilvum_PYR-GCK RHLVVVVGKHREQVAPAALQIGEHLGRTVDIAVQEEQRGTGHAVECGLAA
Mvan_4782|M.vanbaalenii_PYR-1 RHLVVVVGKDRERVAPAALEIGQTLGRDVDIAIQDQQRGTGHAVECGLTA
TH_0911|M.thermoresistible__bu QHLIVVLGHHRDRISPAVEELAATLNRRIDIAVQDRQLGTGHATECGLGA
MMAR_4467|M.marinum_M QHLVVVLGHCHERISPLVDELAESLGRTINVTLQDRPLGTGDAVRCGLSA
MUL_4637|M.ulcerans_Agy99 QHLVVVLGHCHERISPLVDELAESLGRTINVTLQDRPLGTGDAVRCGLSA
Mb1046c|M.bovis_AF2122/97 QRLIVVLGHDHQRIAPLVGELADTLGRTIDVALQDRPLGTGHAVLCGLSA
Rv1018c|M.tuberculosis_H37Rv QRLIVVLGHDHQRIAPLVGELADTLGRTIDVALQDRPLGTGHAVLCGLSA
MAV_1157|M.avium_104 QHLAVVLGHDHERIAPLIADWADDLGRPIDVALQERPRGTGDAVRCGLSA
MLBr_00249|M.leprae_Br4923 QHLVVVLGHEHQRIAPLVAELADTLERTIDVALQDRPRGTGHAVFCGLSA
.:* *:*: :::::. . * * :::::*:. ***.*. *** *
MAB_1148c|M.abscessus_ATCC_199 LPAGFAGTVVVTSGDIPLLDGGTLAGLIGSHTSEPAAATLLTTTLTDPTG
MSMEG_5426|M.smegmatis_MC2_155 LPHDFAGMVVVTSGDVPLLDTATLTGLITSHGSGDAAATVLTTTLPDPTG
Mflv_1944|M.gilvum_PYR-GCK LPADFHGVVVVTAGDVPLLDTDTLAQLIATHDAESADVTLLTTTMADPTG
Mvan_4782|M.vanbaalenii_PYR-1 LPADFTGTVVVTAGDVPLLDTHTLSDLIAAHDAESAAATLLTTTLADPTG
TH_0911|M.thermoresistible__bu LPGDFTGTVVVTSGDVPLLDAATLSELVAAHTATPTAATVLTTTLGDPTG
MMAR_4467|M.marinum_M LPADYSGIVVVTAGDTPLLDAETLADLIETHSATSSAVTVLTTTLSDPHG
MUL_4637|M.ulcerans_Agy99 LPADYSGIVVVTAGDTPLLDAETVADLIETHSATSSAVTVLTTTLSDPHG
Mb1046c|M.bovis_AF2122/97 LPDDYAGNVVVTSGDTPLLDADTLADLIATHRAVSAAVTVLTTTLDDPFG
Rv1018c|M.tuberculosis_H37Rv LPDDYAGNVVVTSGDTPLLDADTLADLIATHRAVSAAVTVLTTTLDDPFG
MAV_1157|M.avium_104 LPDDYAGVLVVTSGDTPLLDADTVADLIAGHTATRAAVTVLTTTLSDPSG
MLBr_00249|M.leprae_Br4923 LPDDYGGIVVVTSGDTPLLDANTLAELIATHNATSAAVTVLTTTFSDPLG
** .: * :***:** **** *:: *: * : : .*:****: ** *
MAB_1148c|M.abscessus_ATCC_199 YGRILRTQDREVIAIVEQTDATESQRAIGEVNAGVYAFDIEPLHAALSRL
MSMEG_5426|M.smegmatis_MC2_155 YGRILRTQDHEIIGIVEQADATESQRTICEVNTGVYAFDIADLRSALTRL
Mflv_1944|M.gilvum_PYR-GCK YGRILRTQDGEVIGIVEQADATESQRAITEVNAAVYAFGVGALRSALSRL
Mvan_4782|M.vanbaalenii_PYR-1 YGRILRTQDGGVIGIVEQADATPSQRAITEVNAAVYAFDAGALRSALSRL
TH_0911|M.thermoresistible__bu YGRVLRTQDGEVIGIVEQADATESQRQIQEVNAGVYAFDIAALRSALSRL
MMAR_4467|M.marinum_M YGRILRTQDNAVMAIVEQTDATPSQREIREVNAGVYAFEITALQSALSRL
MUL_4637|M.ulcerans_Agy99 YGRILRTQDNAVMAIVEQTDATPSQREIREVNAGVYAFEITALQSALSRL
Mb1046c|M.bovis_AF2122/97 YGRILRTQDHGVMAIVEQTDATPSQREIREVNAGVYAFDIAALRSALSRL
Rv1018c|M.tuberculosis_H37Rv YGRILRTQDHEVMAIVEQTDATPSQREIREVNAGVYAFDIAALRSALSRL
MAV_1157|M.avium_104 YGRILRTQDNEVTAIVEHADATESQREIREVNAGVYAFDTAALRSALNRL
MLBr_00249|M.leprae_Br4923 YGRILRTQDNEVMAIIEHADASPSQREIREVNAGVYAFDITALRSALIRL
***:***** : .*:*::**: *** * ***:.**** *::** **
MAB_1148c|M.abscessus_ATCC_199 RSNNAQHELYLTDAVSIIRESGKVVHANHVDDSALVAGVNDRVQLSELGA
MSMEG_5426|M.smegmatis_MC2_155 RSDNAQHELYLTDVVEILRQDHRTVRALHVDDSALVTGVNDRVQLSDLGK
Mflv_1944|M.gilvum_PYR-GCK KTDNAQGELYLTDAIALVRGDGGTVRAVHVDDPALVAGVNDRVQLADLGA
Mvan_4782|M.vanbaalenii_PYR-1 QADNAQGELYLTDVIAIARSDGGVVRARHIDDAALVAGVNDRVQLADLAA
TH_0911|M.thermoresistible__bu HTDNAQQELYLTDVISILRSDGRTVRANHVDDATLVAGVNDRVQLSALAA
MMAR_4467|M.marinum_M SSDNAQQELYLPDVIAILRRDGQTVSARHIDDSALVAGVNDRVQLAQLGA
MUL_4637|M.ulcerans_Agy99 SSDNAQQELYLPDVIAILRRDGQTVSARHIDDSALVAGVNDRVQLAQLGA
Mb1046c|M.bovis_AF2122/97 SSNNAQQELYLTDVIAILRSDGQTVHASHVDDSALVAGVNNRVQLAQLAS
Rv1018c|M.tuberculosis_H37Rv SSNNAQQELYLTDVIAILRSDGQTVHASHVDDSALVAGVNNRVQLAELAS
MAV_1157|M.avium_104 SADNAQQELYLTDVIAILRGDGLPIRARHVDDSALVAGVNNRVQLAQLGA
MLBr_00249|M.leprae_Br4923 NSNNTQQELYLTDVISILRREGQKVNAQHIDDNALVAGVNNRVQLAELSA
::*:* ****.*.: : * . : * *:** :**:***:****: *.
MAB_1148c|M.abscessus_ATCC_199 ELNRRIIRHHQRNGVTIIDPSSTWIDVDVHIGQDATIAPGTQLHSATVIG
MSMEG_5426|M.smegmatis_MC2_155 VLNRRIVAAHQRAGVTIIDPGSTWIDIDVQIGQDTVVHPGTQLLGATRVG
Mflv_1944|M.gilvum_PYR-GCK ELNRRVVAAHQRAGVTIVDPATTWIDVDVTIGRDTVVRPGTQLLGATAVG
Mvan_4782|M.vanbaalenii_PYR-1 VLNRRIVEGHQRAGVTIIDPASTWIDVDVTIGRDTVVRPGTQLLGATTIG
TH_0911|M.thermoresistible__bu ELNRRIIKSHQLAGVTIIDPATTWIDVDVTIGRDTVIRPGTQLLDGTSIG
MMAR_4467|M.marinum_M ELNRRIVAAHQMAGVTVIDPATTWIDVDVAIGRDTVIQPGTQLLGHTQIG
MUL_4637|M.ulcerans_Agy99 ELNRRIVAAHQMAGVTVIDPATTWIDVDAAIGRDTVIQPGTQLLGHTQIG
Mb1046c|M.bovis_AF2122/97 ELNRRVVAAHQLAGVTVVDPATTWIDVDVTIGRDTVIHPGTQLLGRTQIG
Rv1018c|M.tuberculosis_H37Rv ELNRRVVAAHQLAGVTVVDPATTWIDVDVTIGRDTVIHPGTQLLGRTQIG
MAV_1157|M.avium_104 ELNRRIVAAHQLAGVTVVDPATTWIDVDVTIGRDTVIHPGTQLLGRTQIG
MLBr_00249|M.leprae_Br4923 ELNRRIVATHQVAGVTIIDPATTWIDIDVTIGNDTVIHPGTQLLGRTQIG
****:: ** ***::**.:****:*. **.*:.: ***** . * :*
MAB_1148c|M.abscessus_ATCC_199 GHCHIGPDTTLIDVTVGDSATVIRTHGQGSTIGARSVIGPFTYLRPGTVT
MSMEG_5426|M.smegmatis_MC2_155 SHCVIGPDTTLTHVTVGDGASVVRTHGSESVIGAGATVGPFTYLRPGTNL
Mflv_1944|M.gilvum_PYR-GCK GRCEIGPDTTLTDVTVGDGAQVIRTHGTSSQIGAGAVVGPFTYLRPGTAL
Mvan_4782|M.vanbaalenii_PYR-1 GRAEIGPDTTLADVTVGDGAAVIRTHGTSAVIGDDSVVGPFTYLRPGTEL
TH_0911|M.thermoresistible__bu EHCDIGPDTTLSDVAVADRATVCRTHGTASQIGPGATVGPFSYLRPGTVL
MMAR_4467|M.marinum_M DRCEIGPDTTLTDVTVGDNASVVRTHGSSSSIGAAAAVGPFTFLRPGTVL
MUL_4637|M.ulcerans_Agy99 DRCEIGPDTTLTDVTVGDNASVVRTHGSSSSIGAAAAVGPFTYLRPGTVL
Mb1046c|M.bovis_AF2122/97 GRCVVGPDTTLTDVAVGDGASVVRTHGSSSSIGDGAAVGPFTYLRPGTAL
Rv1018c|M.tuberculosis_H37Rv GRCVVGPDTTLTDVAVGDGASVVRTHGSSSSIGDGAAVGPFTYLRPGTAL
MAV_1157|M.avium_104 GHCVVGPDTTLTDVSVGDGASVVRTHGTGSSVGAGATVGPFAYLRPGTVL
MLBr_00249|M.leprae_Br4923 ECCVIGPDTTLTDVLVSQRATVVRTHGTSSTIGAGAMVGPFTYLRPGTVL
. :****** .* *.: * * **** : :* : :***::*****
MAB_1148c|M.abscessus_ATCC_199 GESAKLGAFVEVKNSTVGRGTKVPHLTYVGDADIGEHSNIGASSVFVNYD
MSMEG_5426|M.smegmatis_MC2_155 GADGKLGAFVETKNCTIGTGTKVPHLTYVGDADIGEYSNIGASSVFVNYD
Mflv_1944|M.gilvum_PYR-GCK GADGKLGAFVETKNATIGTGTKVPHLTYVGDADIGEHSNIGASSVFVNYD
Mvan_4782|M.vanbaalenii_PYR-1 GAAGKLGAFVETKNATIGTGTKVPHLTYVGDADIGEHSNIGASSVFVNYD
TH_0911|M.thermoresistible__bu GADGKLGAFVETKNSTIGTGSKVPHLTYVGDADIGEHSNIGASSVFVNYN
MMAR_4467|M.marinum_M GTGGKLGAFVETKNSTIGAGTKVPHLTYVGDADIGDDSNIGAGSVFVNYD
MUL_4637|M.ulcerans_Agy99 GTGGKLGAFVETKNSTIGAGTKVPHLTYVGDADIGDDSNIGAGSVFVNYD
Mb1046c|M.bovis_AF2122/97 GADGKLGAFVEVKNSTIGTGTKVPHLTYVGDADIGEYSNIGASSVFVNYD
Rv1018c|M.tuberculosis_H37Rv GADGKLGAFVEVKNSTIGTGTKVPHLTYVGDADIGEYSNIGASSVFVNYD
MAV_1157|M.avium_104 GDDGKLGAFVETKNATIGTGTKVPHLTYVGDADIGEHSNIGASSVFVNYD
MLBr_00249|M.leprae_Br4923 GTKGKLGAFVETKNSTIGTGAKVPHLTYVGDADIGEHSNIGASSVFVNYD
* .*******.**.*:* *:**************: *****.******:
MAB_1148c|M.abscessus_ATCC_199 GETKRRTVIGSHVKTGSDTMFVAPVTVGDGAYTGAGTVIREDVPPGALAV
MSMEG_5426|M.smegmatis_MC2_155 GENKSRTTIGSHVRTGSDTMFVAPVTVGDGAYTGAGTVLRDDVPPGALAV
Mflv_1944|M.gilvum_PYR-GCK GETKSRTTIGSHVRTGSDTMFVAPVTVGDGAYTGAGTVIRDDVPPGALAV
Mvan_4782|M.vanbaalenii_PYR-1 GETKSRTTIGSHVRTGSDTMFVAPVTVGDGAYTGAGTVVREDVPPGALAV
TH_0911|M.thermoresistible__bu GETKSRTRIGSHVRTGSDTMFIAPVTVGDGAYTGAGTVLREDVPPGALAV
MMAR_4467|M.marinum_M GMTKNRATIGSHVRSGAGTRFVAPVNVGDGAYTGAGTVIRDDVPPGALAV
MUL_4637|M.ulcerans_Agy99 GMTKNRATIGSHVRSGAGTRFVAPVNVGDGAYTGAGTVIRDDVPPGALAV
Mb1046c|M.bovis_AF2122/97 GTSKRRTTVGSHVRTGSDTMFVAPVTIGDGAYTGAGTVVREDVPPGALAV
Rv1018c|M.tuberculosis_H37Rv GTSKRRTTVGSHVRTGSDTMFVAPVTIGDGAYTGAGTVVREDVPPGALAV
MAV_1157|M.avium_104 GESKRRTTVGSHVRTGSDTMFVAPVTVGDGAYTGAGTVVREDVPPGALAV
MLBr_00249|M.leprae_Br4923 GTAKQRTTIGSHVRTGSDTKFVAPVTVGDGAYTGAGTVVRNDVPPGALAV
* * *: :****::*:.* *:***.:***********:*:*********
MAB_1148c|M.abscessus_ATCC_199 SAGRQRNIEGWVAQKRPGSDAAKAAEEASKGS------------------
MSMEG_5426|M.smegmatis_MC2_155 SAGPQRNIEGWVAKKRPGSAADKAARKALGDES-----------------
Mflv_1944|M.gilvum_PYR-GCK SAGPQRNIEGWVARKRPGSKAAAAAEAAAADGDTAAADRAAATAKPAPAT
Mvan_4782|M.vanbaalenii_PYR-1 SAGPQRNIEGWVTRKRPGSAAAEAAAAAGAGAGAAAEDQGPE------AT
TH_0911|M.thermoresistible__bu SAGPQRTIEGWVARKRPGSKSDLAAKAAAERDGDDTPKTS----------
MMAR_4467|M.marinum_M SGGPQRNIEDWVQQKRPGTPSAEAARKASAEQSTPPPDADQTP-------
MUL_4637|M.ulcerans_Agy99 SGGPQRNIEDWVQQKRPGTPSAEAARKASAEQSTPPPDADHPP-------
Mb1046c|M.bovis_AF2122/97 SAGPQRNIENWVQRKRPGSPAAQASKRASEMACQQPTQPPDADQTP----
Rv1018c|M.tuberculosis_H37Rv SAGPQRNIENWVQRKRPGSPAAQASKRASEMACQQPTQPPDADQTP----
MAV_1157|M.avium_104 SAGPQRNIEGWVRRKRPGSAAARAAEAAEKAAGGRPAGEAE---------
MLBr_00249|M.leprae_Br4923 SVSPQRNIENWVQRKRPGSAAAQAAEKASTRTGKQSQQKSEPD-------
* . **.**.** :****: : *: *
MAB_1148c|M.abscessus_ATCC_199 --
MSMEG_5426|M.smegmatis_MC2_155 --
Mflv_1944|M.gilvum_PYR-GCK GE
Mvan_4782|M.vanbaalenii_PYR-1 GE
TH_0911|M.thermoresistible__bu --
MMAR_4467|M.marinum_M --
MUL_4637|M.ulcerans_Agy99 --
Mb1046c|M.bovis_AF2122/97 --
Rv1018c|M.tuberculosis_H37Rv --
MAV_1157|M.avium_104 --
MLBr_00249|M.leprae_Br4923 --