For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MEARKAPNPLEPYVVISADCHAELPTEQYREYVDPDYREDFEVYLAEKAAASQVGGFIDEQFAQDWFSEH GEGIAGGWDVGLRDKELDSDGVVGEVIFPDADAVSGVAGAPFGAGLAQSGSLDPGRAMAGARAHNRWLAE LCSHSPERRAGVAVVPILADVDAAVAEITRAAESGLRGGIMIPALWASYPPYHDRRYDKVWAACQDLQMP VHTHVGAAPQQEYGEHLGIYVTEVRWWGVRPLWFALWSGVFERFPGLRWGATECGAFWANDLLWLMDTRY LREHSAKKMSHLLEGDLTMPPSAYFDRNCFIGATTTERRELARRYEIGVSNMLWGNDFPHPEGTWPHTRE WLRRSFWDIPIDETRQILGLAAAEVYNFDLDALAPLAGRIGPTPADLGQDDAVSVPKWEAARQAGRHWLT GAEPLADLVQN
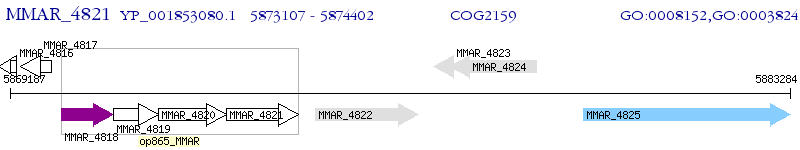
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4821 | - | - | 100% (431) | hypothetical protein MMAR_4821 |
| M. marinum M | MMAR_4820 | - | 7e-74 | 39.85% (399) | hypothetical protein MMAR_4820 |
| M. marinum M | MMAR_2618 | - | 1e-21 | 27.07% (399) | amidohydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3540c | - | 6e-05 | 31.93% (119) | hypothetical protein Mb3540c |
| M. gilvum PYR-GCK | Mflv_2441 | - | 6e-19 | 24.17% (393) | amidohydrolase 2 |
| M. tuberculosis H37Rv | Rv3510c | - | 6e-05 | 31.93% (119) | hypothetical protein Rv3510c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_3540 | - | 0.0 | 89.34% (422) | amidohydrolase |
| M. smegmatis MC2 155 | MSMEG_4837 | - | 3e-20 | 24.81% (399) | amidohydrolase 2 |
| M. thermoresistible (build 8) | TH_2339 | - | 8e-13 | 23.18% (371) | amidohydrolase 2 |
| M. ulcerans Agy99 | MUL_0389 | - | 3e-71 | 39.35% (399) | hypothetical protein MUL_0389 |
| M. vanbaalenii PYR-1 | Mvan_2022 | - | 5e-18 | 24.17% (393) | amidohydrolase 2 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4821|M.marinum_M MEARKAPNPLEPYVVISADCHAELPTEQYREYVDPDYREDFEVYLAEKAA
MAV_3540|M.avium_104 ------MTDVDPYIIISADTHAELPTERYREYVDPEYREDFETYLAEKTA
MUL_0389|M.ulcerans_Agy99 ---------MDRYTVISADCHAGADLFDYREYLEAKYQDEFDDWA--KTF
Mflv_2441|M.gilvum_PYR-GCK -------MDKNDMILISVDDHIVEPPDMFKNHLSKKYLDEAPRLVHNEDG
Mvan_2022|M.vanbaalenii_PYR-1 -------MNRDDMILISVDDHIVEPPDMFKNHLPSKYLDEAPRLVHNPDG
MSMEG_4837|M.smegmatis_MC2_155 -------MNKDDMILISVDDHIIEPPDMFKNHLPAKYRDEAPRLVHNPDG
TH_2339|M.thermoresistible__bu ---MSHDLRPEDLILVSIDDHVVEPPDMFLRHVPAKYRDEAPIVVTDENG
Mb3540c|M.bovis_AF2122/97 ---------------MTIDVWMQHPT---QRFLHGDMFASLRRWTG----
Rv3510c|M.tuberculosis_H37Rv ---------------MTIDVWMQHPT---QRFLHGDMFASLRRWTG----
:: * ..: . .
MMAR_4821|M.marinum_M AS--QVGGFIDEQFAQDWFSEHGEGIAGGWDVGLRDKELDSDGVVGEVIF
MAV_3540|M.avium_104 AA--QAGGFIDEEFAQEWFSEHGEGIAGGWDVALRDKELDGDGVVGEVIF
MUL_0389|M.ulcerans_Agy99 VN--PFGDLSEPDAEHNWDSDR------------RNAELDSDGVAGEVIF
Mflv_2441|M.gilvum_PYR-GCK SDTWQFRDVVIPNVALNAVAGRPKEEYGLEPQGLDEIRPGCWQVDERVKD
Mvan_2022|M.vanbaalenii_PYR-1 SDTWQFRDVVIPNVALNAVAGRPKEEYGLEPQGLDEIRPGCWQVDERVKD
MSMEG_4837|M.smegmatis_MC2_155 SDTWQFRDTVIPNVALNAVAGRPKEEYGLEPQGLDEIRKGCYDPNERVKD
TH_2339|M.thermoresistible__bu VDQWMYQGRPQGVSGLNAVVSWPPEEWGRNPARFAEMRPGVYDVHERVRD
Mb3540c|M.bovis_AF2122/97 ----------------------------------GSIPETDIPIEATVSS
Rv3510c|M.tuberculosis_H37Rv ----------------------------------GSIPETDIPIEATVSS
. *
MMAR_4821|M.marinum_M PDADAVSGVAGAPFGAGLAQ--SGSLDPGRAMAGARAHNRWLAELCSHSP
MAV_3540|M.avium_104 PDADAVTGVAGAPFGAGLGQ--SGDLDPGRAMAGARAHNRWLAELCSHSP
MUL_0389|M.ulcerans_Agy99 PNTVPPFFPQGSLAAPAPST--ARDLELR--WAGLRAHNRWLADFCSLSP
Mflv_2441|M.gilvum_PYR-GCK MNAGGILGSMCFPSFPGFAGRLFATEDPEFSLALVQAYNDWHVEEWCGAY
Mvan_2022|M.vanbaalenii_PYR-1 MNAGGILGSMCFPSFPGFAGRLFATEDAEFSLALVQAYNDWHVEEWCGAY
MSMEG_4837|M.smegmatis_MC2_155 MNAGGVLATMNFPSFPGFAARLFATDDEEFSLALVQAYNDWHIDEWCASN
TH_2339|M.thermoresistible__bu MNRNGILASMCFPTFTGFSARHLNMHREHVTLIMVSAYNDWHIDEWAGSY
Mb3540c|M.bovis_AF2122/97 MDAGGVT---------------LGLLSAWRGPNGQDLISNDAVAEWVRLY
Rv3510c|M.tuberculosis_H37Rv MDAGGVT---------------LGLLSAWRGPNGQDLISNDAVAEWVRLY
: .
MMAR_4821|M.marinum_M ERRAGVAVVPILADVDAAVAEITRAAESGLRGGIMIP--ALWASYPPYHD
MAV_3540|M.avium_104 ERRAGVAVVPILADVDAAVAEITRAAESGLRGGILIP--ARWVGYPPYHD
MUL_0389|M.ulcerans_Agy99 ERRAGVGQI-LLGDVDEAVAEVAQIAKLGLPGGVLLPGVAPGTGIPALYA
Mflv_2441|M.gilvum_PYR-GCK PARFIPMTLPVIWDPEACAKEIRRNAARGVHS-LTFSENPSAMGYPSFHD
Mvan_2022|M.vanbaalenii_PYR-1 PARFIPMTLPVIWDPVACAAEIRRNAARGVHS-LTFSENPSAMGYPSFHD
MSMEG_4837|M.smegmatis_MC2_155 PGRFIPMAIPAIWNPGLAAAEVRRVAEKGVHS-LTFTENPSTLGYPSFHD
TH_2339|M.thermoresistible__bu PGRFIPIAILPTWNPEAMCAEIRRVAAKGCRA-VTMPELPHLEGLPSYHD
Mb3540c|M.bovis_AF2122/97 PNRFAGLAAVDLDRPMAAVRELRRRVGEGFVG---LRVVPWLWGAPP--T
Rv3510c|M.tuberculosis_H37Rv PNRFAGLAAVDLDRPMAAVRELRRRVGEGFVG---LRVVPWLWGAPP--T
* *: : . * . : . . *.
MMAR_4821|M.marinum_M RR-YDKVWAACQDLQMPVHTHVG---AAPQQEYGEHLGIYVTEVRWWGVR
MAV_3540|M.avium_104 RR-YDKVWAACQDLQMPVHTHSG---PAPQEEYGGHLGIYVTEVRWWGAR
MUL_0389|M.ulcerans_Agy99 EH-WEPLWAACAQAGLVINHHGGNAGPSPIDGWGSSLAVLVYESHWWAHR
Mflv_2441|M.gilvum_PYR-GCK FDHWKPMWDALVDTDTVLNVHIGSSGRLAITAPDAPMDVMITLQPMNIVQ
Mvan_2022|M.vanbaalenii_PYR-1 FEHWKPMWDALVDTDTVLNVHIGSSGRLAITAPDAPMDVMITLQPMNIVQ
MSMEG_4837|M.smegmatis_MC2_155 LEYWRPLWQALVDTETVMNVHIGSSGKLAITAPDAPMDVMITLQPMNIVQ
TH_2339|M.thermoresistible__bu EDYWGPVFRTLSEENVVMCLHIGTGFGAISMAPDAPIDNMIILATQVSAM
Mb3540c|M.bovis_AF2122/97 DRRYYPLFAECVQSAVPFCTQVGHTGPLRPSETGRPIP------------
Rv3510c|M.tuberculosis_H37Rv DRRYYPLFAECVQSAVPFCTQVGHTGPLRPSETGRPIP------------
: :: : . : * . :
MMAR_4821|M.marinum_M PLWFALWSGVFERFPGLRWGATECGAFWAN---DLLWLMDTRYLREHSAK
MAV_3540|M.avium_104 PLWFALWSGVFERFPGLRWGVTECGAFWAN---DLLWLMDTRYLREHSAK
MUL_0389|M.ulcerans_Agy99 ALLHLIFSGVLDRYPDLTVVLTEQGTGWIPATLDSLDVAAARYRRPNSAI
Mflv_2441|M.gilvum_PYR-GCK AAADLLWSRPIKEYPDLKIALSEGGTGWIP---YFLERVDRTYEMHSTWT
Mvan_2022|M.vanbaalenii_PYR-1 AAADLLWSRPIKEYPDLKIALSEGGTGWIP---YFLERVDRTYEMHSTWT
MSMEG_4837|M.smegmatis_MC2_155 AAADLLWSAPIKEFPTLKIALSEGGTGWIP---YFLDRVDRTFEMHSTWT
TH_2339|M.thermoresistible__bu CAQDLLWGPAMRNYPDLKFAFSEGGIGWIP---FYLDRCDRHYTN-QKWL
Mb3540c|M.bovis_AF2122/97 -----YIDQVALDFPELVIVCGHVGYPWTE----EMVAVARKHEN-----
Rv3510c|M.tuberculosis_H37Rv -----YIDQVALDFPELVIVCGHVGYPWTE----EMVAVARKHEN-----
. :* * . * * : .
MMAR_4821|M.marinum_M K-MSHLLEGDLTMPPSAYFDRNCFIGATTTERRELARRYEIGVSNMLWGN
MAV_3540|M.avium_104 K-MSHLMEGDLTMPPSAYFDRNCFIGATTTERRELARRYEIGVSNILWGN
MUL_0389|M.ulcerans_Agy99 ARFAGPTAGSLPLQPSQYWARQCYVGASFLRPVECAQRHRIGVDRIMWAS
Mflv_2441|M.gilvum_PYR-GCK G-------QDFKGKLPSEVFREHFLTCFIADPVGVATRHQIGVDNICWEA
Mvan_2022|M.vanbaalenii_PYR-1 G-------QDFKGKLPSEVFREHFLTCFIADPVGVATRHQIGVDNICWEA
MSMEG_4837|M.smegmatis_MC2_155 H-------QNFGGKLPSEVFREHFMTCFISDPVGVKNRHMIGIDNICWEM
TH_2339|M.thermoresistible__bu R-------RDFGDKLPSDVFREHSLACYVTDKTSLKLRHEIGIDIIAWEC
Mb3540c|M.bovis_AF2122/97 ----------------VYIDTSAYTIKRLPGKLVRFMKTDTGQRKVLFGT
Rv3510c|M.tuberculosis_H37Rv ----------------VYIDTSAYTIKRLPGKLVRFMKTDTGQRKVLFGT
. : * : :
MMAR_4821|M.marinum_M DFPHPEGTWPHTREWLRRSFWDIPID--ETRQILGLAAAEVYNFDLDALA
MAV_3540|M.avium_104 DFPHPEGTWPHTRDWLRRSFWDIPVE--ETRQMLGLAAAELYNFDLGALA
MUL_0389|M.ulcerans_Agy99 DYPHMEGTAPYSREALRHTFSDVDPD--EVAAMVGGNAAAVYGFDLQALA
Mflv_2441|M.gilvum_PYR-GCK DYPHSDSMWPGAPEQLDEVLKANNVPDDEINKMTYENAMRWYHWDPFTHI
Mvan_2022|M.vanbaalenii_PYR-1 DYPHSDSMWPGAPEQLDEVLKVNNVPDDEIDKMTYQNAMRWYHWDPFTHI
MSMEG_4837|M.smegmatis_MC2_155 DYPHSDSMWPGAPEELSAVFDTYDVPDDEINKITHENAMRLYHFDPFAHV
TH_2339|M.thermoresistible__bu DYPHSDCFWPDAPEQVHAELRAAGADDFDINKITWENSCRFFSWDPFEHT
Mb3540c|M.bovis_AF2122/97 NYPMIAHTHALT------GLDELGLSDEARRDFLHGNAVRVFKLDPRGKV
Rv3510c|M.tuberculosis_H37Rv NYPMIAHTHALT------GLDELGLSDEARRDFLHGNAVRVFKLDPRGKV
::* . : : : : : *
MMAR_4821|M.marinum_M PLAGRIGPTPADLGQDDAVSVPKWEAARQAGRHW----LTGAEPLADLVQ
MAV_3540|M.avium_104 PLAARIGPVPADLGQDDAVSIPKWEAARRTGRHW----LTDAEPMPDLVQ
MUL_0389|M.ulcerans_Agy99 PLAARIGPTVTEVAEP-LAAIPADASS------------TAFEPDPIRAW
Mflv_2441|M.gilvum_PYR-GCK AKEQATVGALRKAAEGHDVSIQALSNK---EKTG--TTFSDFAASAKSIS
Mvan_2022|M.vanbaalenii_PYR-1 PKEQATVGALRKAAEGHDVSIQALSHHKEGERGGGAGHFDALNAAARGNS
MSMEG_4837|M.smegmatis_MC2_155 PKEQATVGALRKAAEGHDVSIRALSKK---DKTG--ASFADFQANAKAVS
TH_2339|M.thermoresistible__bu PRAEATVGALRAKATDVDTTIRSRAEW------------AQLYEQKQQFA
Mb3540c|M.bovis_AF2122/97 QT------------------------------------------------
Rv3510c|M.tuberculosis_H37Rv QT------------------------------------------------
MMAR_4821|M.marinum_M N---
MAV_3540|M.avium_104 S---
MUL_0389|M.ulcerans_Agy99 ----
Mflv_2441|M.gilvum_PYR-GCK GNKD
Mvan_2022|M.vanbaalenii_PYR-1 GAD-
MSMEG_4837|M.smegmatis_MC2_155 GARD
TH_2339|M.thermoresistible__bu QA--
Mb3540c|M.bovis_AF2122/97 ----
Rv3510c|M.tuberculosis_H37Rv ----