For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNTALDITTNGPIRTWTINLPDVGNAITGKDFIAAFEAAVDDANIDTSVRAVILTGAGKIFSGGGNVKEM ANRDGMFGLDALEQRQAYTDGIQRIPRALARLEVPLIGAINGAAIGAGCDLAMMCDIRIASQRASFAESF VALGIIPGDGGTWFLQRAIGYERAAEMTLTGDRIDAATALEWGMVSRVVPHDDLLPQAQELAERIVKNPA HALRMAKRLLQESRVGSLESTLGMAAAMQPLAHRDPEHQQRIAKWRTR
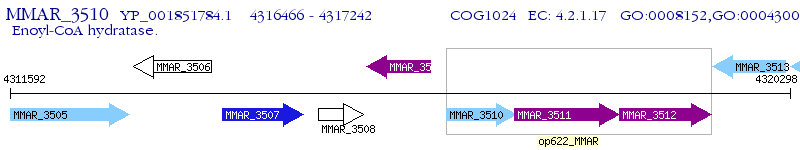
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3510 | echA8_5 | - | 100% (258) | enoyl-CoA hydratase, EchA8_5 |
| M. marinum M | MMAR_4396 | echA8 | 2e-32 | 37.96% (216) | enoyl-CoA hydratase EchA8 |
| M. marinum M | MMAR_1903 | echA16_2 | 7e-28 | 37.04% (216) | enoyl-CoA hydratase, EchA16_2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1099c | echA8 | 9e-34 | 38.89% (216) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_4495 | - | 1e-31 | 34.89% (235) | enoyl-CoA hydratase |
| M. tuberculosis H37Rv | Rv1070c | echA8 | 9e-34 | 38.89% (216) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 2e-34 | 39.35% (216) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_1185c | - | 2e-30 | 36.24% (218) | enoyl-CoA hydratase |
| M. avium 104 | MAV_1884 | - | 8e-34 | 33.33% (258) | enoyl-CoA isomerase |
| M. smegmatis MC2 155 | MSMEG_2880 | - | 1e-112 | 78.88% (251) | enoyl-CoA hydratase |
| M. thermoresistible (build 8) | TH_3045 | - | 2e-38 | 37.40% (254) | PUTATIVE Enoyl-CoA hydratase/isomerase |
| M. ulcerans Agy99 | MUL_2758 | echA8_5 | 1e-145 | 99.22% (258) | enoyl-CoA hydratase, EchA8_5 |
| M. vanbaalenii PYR-1 | Mvan_2877 | - | 1e-115 | 80.63% (253) | enoyl-CoA hydratase/isomerase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3510|M.marinum_M ---------------MNTALDITTNGPIRTWTINLPDVGNAITGKDFIAA
MUL_2758|M.ulcerans_Agy99 ---------------MNTALDITTNGPIRTWTINLPDVGNAITGRDFIAA
MSMEG_2880|M.smegmatis_MC2_155 -------------MTASEPLLIGASGAAQMWTINLPQVGNAITGSDFIRA
Mvan_2877|M.vanbaalenii_PYR-1 MTPTSPHRSERQPLDTASPLAVHTVDAVTTWTINLPHVGNAITGTDFIAA
TH_3045|M.thermoresistible__bu -----------VKYDLPDVMTVRADGPIRTVTLDRAADLNSVD-ADLHWA
MAV_1884|M.avium_104 ---------------MNPGISIEREGGILRLTLDRPDKLNAVD-TPMLNE
Mb1099c|M.bovis_AF2122/97 --------------MTYETILVERDQRVGIITLNRPQALNALN-SQVMNE
Rv1070c|M.tuberculosis_H37Rv --------------MTYETILVERDQRVGIITLNRPQALNALN-SQVMNE
MLBr_02402|M.leprae_Br4923 --------------MKYDTILVDGYQRVGIITLNRPQALNALN-SQMMNE
MAB_1185c|M.abscessus_ATCC_199 -----------MTSHNFETILTERIDRVAVITLNRPKALNALN-SQVMNE
Mflv_4495|M.gilvum_PYR-GCK --------------MTEDLVLTADHDGVRVITLNRPAARNALS-RDLIRA
: *:: . *:: .
MMAR_3510|M.marinum_M FEAAVDDANIDTSVRAVILTGAGKIFSGGGNVKEMANRDGMFGLDALEQR
MUL_2758|M.ulcerans_Agy99 FEAAVDDANIDTSVRAVILTGAGKIFSGGGNVKEMANRDGMFGLDALEQR
MSMEG_2880|M.smegmatis_MC2_155 FEDAVDAANRDPTVGAVILTGSGKIFSAGGNVKEMADREGMFGLDAIDQR
Mvan_2877|M.vanbaalenii_PYR-1 FESAVDTANSDVAVRAVILTGAGKIFSAGGNVKEMADRQGMFGLDAIAQR
TH_3045|M.thermoresistible__bu LANVWRQLAADTDARVVILTGAGRAFSAGGDLDWITS----FLTDQVARD
MAV_1884|M.avium_104 LLGRLDESADDASVRAVLLTGAGRAFCSGGDLTGGDT-------------
Mb1099c|M.bovis_AF2122/97 VTSAATELDDDPDIGAIIITGSAKAFAAGADIKEMAD---------LTFA
Rv1070c|M.tuberculosis_H37Rv VTSAATELDDDPDIGAIIITGSAKAFAAGADIKEMAD---------LTFA
MLBr_02402|M.leprae_Br4923 ITNAAKELDIDPDVGAILITGSPKVFAAGADIKEMAS---------LTFT
MAB_1185c|M.abscessus_ATCC_199 VTTAAAEFDADHGIGAIIITGSEKAFAAGADIKEMSE---------QSFS
Mflv_4495|M.gilvum_PYR-GCK TYTALTSADDDDAVRAVVLTGADPAFCAGVDLKEAAR---------DGLS
* .:::**: *..* ::
MMAR_3510|M.marinum_M QAYTDGIQRIPRALARLEVPLIGAINGAAIGAGCDLAMMCDIRIASQRAS
MUL_2758|M.ulcerans_Agy99 QAYTDGIQRIPRALARLEVPLIGAINGAAIGAGCDLAMMCDIRIASQRAS
MSMEG_2880|M.smegmatis_MC2_155 FAYVDGIQRIPRALARLEVPIIAAVNGAAVGAGFDLAMMCDIRVASERAS
Mvan_2877|M.vanbaalenii_PYR-1 HAYVDGIQRIPRALARLEVPLIAAVNGPAIGAGCDLAAMCDIRVASERAS
TH_3045|M.thermoresistible__bu ESIREGAQIIEEMLR-FPLPVIAAVNGPAVGLGCSVAVLCDIVLIAENAY
MAV_1884|M.avium_104 EGAAQAANDVVRAITELPKPVVAGVHGPAVGVGCALALACDLVVATRAAY
Mb1099c|M.bovis_AF2122/97 DAFTADFFATWGKLAAVRTPTIAAVAGYALGGGCELAMMCDVLIAADTAK
Rv1070c|M.tuberculosis_H37Rv DAFTADFFATWGKLAAVRTPTIAAVAGYALGGGCELAMMCDVLIAADTAK
MLBr_02402|M.leprae_Br4923 DAFDADFFSAWGKLAAVRTPMIAAVAGYALGGGCELAMMCDLLIAADTAK
MAB_1185c|M.abscessus_ATCC_199 DMFGSDFFSAWGKLGAVRTPTIAAVSGYALGGGCELAMMCDVIIAAENAT
Mflv_4495|M.gilvum_PYR-GCK YFAEFRTQSCIAAVAAMRTPVVGAINGATFTGGLEMALGCDFMIASERAV
: . * :..: * :. * :* **. : : *
MMAR_3510|M.marinum_M FAESFVALGIIPGDGGTWFLQRAIGYERAAEMTLTGDRIDAATALEWGMV
MUL_2758|M.ulcerans_Agy99 FAESFVALGIIPGDGGTWFLQRAIGYERAAEMTLTGDRIDAATALEWGMV
MSMEG_2880|M.smegmatis_MC2_155 FAESFVQLGLVPGDGGTWFLQRAIGYERAAEMTFTGDRVDAATALDWGLV
Mvan_2877|M.vanbaalenii_PYR-1 FAESFVQIGLVPGDGGTWFLQRAIGYERAAEMTLTGDRIDAATALEWGLV
TH_3045|M.thermoresistible__bu LADPHVAVGLVAGDGGAAMWPLLTLVLRSREYLFTGDRIPAATAVELGLA
MAV_1884|M.avium_104 FQLAFSKLGLMLDGGASVLLPAAIGRARAARVAMTAEKITAATAFEWGLI
Mb1099c|M.bovis_AF2122/97 FGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDLILTGRTMDAAEAERSGLV
Rv1070c|M.tuberculosis_H37Rv FGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDLILTGRTMDAAEAERSGLV
MLBr_02402|M.leprae_Br4923 FGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDLILTGRTIDAAEAERSGLV
MAB_1185c|M.abscessus_ATCC_199 FGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDMILTGRNMDAAEAERSGLV
Mflv_4495|M.gilvum_PYR-GCK FADTHARVGILPGGGMTARLPQLVGAAMARRLSMTGEVVDAARAERIGLV
: . :*:: . * : : :*. : ** * *:
MMAR_3510|M.marinum_M SRVVPHDDLLPQAQELAERIVKNPAHALRMAKRLLQESRVGSLESTLGMA
MUL_2758|M.ulcerans_Agy99 SRVVPHGDLLPQAQELAERIVKNPAHALRMAKRLLQESRVGSLESTLGMA
MSMEG_2880|M.smegmatis_MC2_155 SRVVPHDELLSAAQDLAARIVRNPARALRMAKRLLQESRTGVLESTLGMA
Mvan_2877|M.vanbaalenii_PYR-1 SRVVAHDELLTAAGELAARIAKNPPHTLRMAKRLLQESRTGVLESTLGMA
TH_3045|M.thermoresistible__bu SRTVPADDLLPEAIRLAERLAAQPAEALRGTKRIVNMYLSQALAGPLQAG
MAV_1884|M.avium_104 SYVVDDDAYETDLAELTSTLANGPTHSYRWIKRALSAATLSDLESVQSLE
Mb1099c|M.bovis_AF2122/97 SRVVPADDLLTEARATATTISQMSASAARMAKEAVNRAFESSLSEGLLYE
Rv1070c|M.tuberculosis_H37Rv SRVVPADDLLTEARATATTISQMSASAARMAKEAVNRAFESSLSEGLLYE
MLBr_02402|M.leprae_Br4923 SRVVLADDLLPEAKAVATTISQMSRSATRMAKEAVNRSFESTLAEGLLHE
MAB_1185c|M.abscessus_ATCC_199 SRVVATESLLDEAKAVAKTISEMSLSASMMAKEAVNRAFESSLAEGLLFE
Mflv_4495|M.gilvum_PYR-GCK TEVVAHQHLLQRAIDLAHQIAEVPRPTMRGLKEIYTAGASAVVDPALAAE
: .* : : . : *. :
MMAR_3510|M.marinum_M AAMQPLAHRDPEHQQRIAKWRTR---------
MUL_2758|M.ulcerans_Agy99 AAMQPLAHRDPEHQQRIAKWRTRIAKWRTR--
MSMEG_2880|M.smegmatis_MC2_155 AAMQPLAHHDGEHARRIEKWRSV---------
Mvan_2877|M.vanbaalenii_PYR-1 AAMQPLAHRDPEHAARISRWRSS---------
TH_3045|M.thermoresistible__bu FAAEVATMRTAEHRDRLLALRERTR-------
MAV_1884|M.avium_104 ADGQRLLVRTSDFAEGVRAFRERRPTEFRGE-
Mb1099c|M.bovis_AF2122/97 RRLFHSAFATEDQSEGMAAFIEKRAPQFTHR-
Rv1070c|M.tuberculosis_H37Rv RRLFHSAFATEDQSEGMAAFIEKRAPQFTHR-
MLBr_02402|M.leprae_Br4923 RRLFHSTFVTDDQSEGMAAFIEKRAPQFTHR-
MAB_1185c|M.abscessus_ATCC_199 RRIFHSAFGTADQSEGMAAFVEKRPANFIHR-
Mflv_4495|M.gilvum_PYR-GCK ERIAFAQHRDLEGLGDRFSAVSQRNRDQIGGA
: