For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTDVLAGADARLEVVSVASGRMRVHVSGFGVDAVRAVAIEEKVAKVTGVQAVQAYPRTASVVIWYSPQH CDTAEVLSAIADAEHVPAESVPARAPHSADVPVTGVVRTVIGGPALALFGLCRRLLGGSPDGDRPGDTVD GGSCGQGAVAACHSSPGQDSGGERRKWLRRVWLTWPLALLATASTMVFGAAPWAGWLAFAATVPVQFVAG WPILKAAAQQARALSANMDTLIALGTLTAFGYSTYQLFAGGPLFFDTAALIIAFVVLGRYFEARATGKTS EAISKLLEMGAKEATLLVDDQELLVPVDQVRVGDLVRVRPGEKIPVDGEVTDGRAAVDESMLTGESVPVE KTVGDRVAGATVNTDGLLTVRATAVGADTALAQIVRLVEQAQGGKAPVQRLADRVSAVFVPVVVGVAAAT FVGWSLIAADPVGGMTAAVAVLIIACPCALGLATPTAITVGTGRGAEMGILVKGGEVLEASKKIDTVVFD KTGTLTRAQMRLTDVIAGKRRQPDPVLRIAAAVESGSEHPIGAAIVAGARERGLEIPSATGFANLAGHGV RAEVNGKPVLVGRRKLVDESDLVLPDHLAAAAAELEQQGRTAVFVGQDDQVVGVLAVADTVKDDAIEVVR RLHAIGLKVAMITGDNARTAAAIAQQVGIDQVLAEVLPADKVTEIRRLQNQGQVVAMVGDGVNDAPALVQ ADLGIAIGTGTDVAIEASDITLMSGQLDGVVRAIGLSRQTLRTIYQNLGWAFGYNTAAIPLAALGLLNPV IAGAAMGLSSVSVVTNSLRLRRFGRDAQQQPERHTAVAPIRT
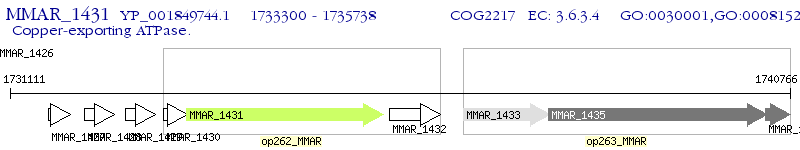
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1431 | zntA | - | 100% (812) | cation transport ATPase, ZntA |
| M. marinum M | MMAR_2536 | - | 0.0 | 81.14% (774) | metal cation transporter p-type ATPase |
| M. marinum M | MMAR_4876 | ctpV | 0.0 | 78.18% (802) | metal cation transporter p-type ATPase, CtpV |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0994 | ctpV | 0.0 | 81.83% (776) | metal cation transporter P-type ATPase CtpV |
| M. gilvum PYR-GCK | Mflv_2244 | - | 0.0 | 53.11% (659) | heavy metal translocating P-type ATPase |
| M. tuberculosis H37Rv | Rv0969 | ctpV | 0.0 | 81.96% (776) | metal cation transporter P-type ATPase CtpV |
| M. leprae Br4923 | MLBr_01987 | ctpA | 1e-146 | 46.74% (629) | putative cation-transporting ATPase |
| M. abscessus ATCC 19977 | MAB_3984c | - | 1e-154 | 45.89% (717) | putative metal transporter ATPase |
| M. avium 104 | MAV_5246 | - | 1e-162 | 48.68% (680) | copper-translocating P-type ATPase |
| M. smegmatis MC2 155 | MSMEG_5014 | - | 0.0 | 53.76% (651) | copper-translocating P-type ATPase |
| M. thermoresistible (build 8) | TH_4361 | - | 1e-178 | 46.79% (780) | PUTATIVE heavy metal translocating P-type ATPase |
| M. ulcerans Agy99 | MUL_0423 | ctpV | 0.0 | 78.59% (780) | metal cation transporter p-type ATPase, CtpV |
| M. vanbaalenii PYR-1 | Mvan_4449 | - | 1e-180 | 53.30% (651) | heavy metal translocating P-type ATPase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0994|M.bovis_AF2122/97 ----------------------MRVCVTGFNVDAVRAVAIEETVSQVTGV
Rv0969|M.tuberculosis_H37Rv ----------------------MRVCVTGFNVDAVRAVAIEETVSQVTGV
MMAR_1431|M.marinum_M MTTDVLAGADARLEVVSVASGRMRVHVSGFGVDAVRAVAIEEKVAKVTGV
MUL_0423|M.ulcerans_Agy99 ----------------------MRVHANGFAIDATRAAAIEESVGRVGGV
Mflv_2244|M.gilvum_PYR-GCK ---------------------------------------MNTLELSIGGM
Mvan_4449|M.vanbaalenii_PYR-1 ---------------------------------------MNTIELSIGGM
MSMEG_5014|M.smegmatis_MC2_155 -----------------------------------MTTATGHVELQIGGM
TH_4361|M.thermoresistible__bu ----------------------------------MNTGGMNTVTLTVGGM
MAV_5246|M.avium_104 --------------------MSTPPRHIDEGTFPDHTASTARIELEITGM
MAB_3984c|M.abscessus_ATCC_199 --------------------------------METTNTGMETTDLRLHGM
MLBr_01987|M.leprae_Br4923 -------------------------------------------------M
:
Mb0994|M.bovis_AF2122/97 HAVHAYPRTASVVIWYSPELGDTAAVLSAITKAQHVPAELVPARAPHSAG
Rv0969|M.tuberculosis_H37Rv HAVHAYPRTASVVIWYSPELGDTAAVLSAITKAQHVPAELVPARAPHSAG
MMAR_1431|M.marinum_M QAVQAYPRTASVVIWYSPQHCDTAEVLSAIADAEHVPAESVPARAPHSAD
MUL_0423|M.ulcerans_Agy99 RAVRAYPRTASVVIWYSPAHCDTAAIVSAIAEAEHAGDASAPAPAARVAG
Mflv_2244|M.gilvum_PYR-GCK TCASCAARVEKKLNKLD----GVTATVNYATEKARVEFGDNVSTDQLVSA
Mvan_4449|M.vanbaalenii_PYR-1 TCASCAARVEKKLNKLD----GVTATVNFATEKARVAFGDEVSPGQLVSA
MSMEG_5014|M.smegmatis_MC2_155 TCASCAARIERKLNKLD----GVTASVNYATEKASVEVAGEVSPETLIEV
TH_4361|M.thermoresistible__bu TCASCAARVEKRLNRID----GVVATVNFATEQATVRYPDHVDPADLIAA
MAV_5246|M.avium_104 TCASCAARIEKKLNKLD----GVTATVNYATEKAAVSAPASYDPQTLITE
MAB_3984c|M.abscessus_ATCC_199 TCASCASRVEKGLNKIT----GVNARVNLALESARVEHDGSVTGEDLVKA
MLBr_01987|M.leprae_Br4923 RAPNGWNNLPNKLSDFS-------TLVNSATRVAAINLSEVTQTAEVCEA
. . : :.
Mb0994|M.bovis_AF2122/97 VRGVGVVRKITGG--------IRRMLSR---------PPGVDKPLKASRC
Rv0969|M.tuberculosis_H37Rv VRGVGVVRKITGG--------IRRMLSR---------PPGVDKPLKASRC
MMAR_1431|M.marinum_M VPVTGVVRTVIGGPALALFGLCRRLLGG---------SPDGDRPGDTVDG
MUL_0423|M.ulcerans_Agy99 IADGGVVGRVIGG-------LARALLGMRTDDNDDDPPPEIARLRAALDR
Mflv_2244|M.gilvum_PYR-GCK VESAGYQAALPEP-------------------------------------
Mvan_4449|M.vanbaalenii_PYR-1 VESAGYQAELPER-------------------------------------
MSMEG_5014|M.smegmatis_MC2_155 VETAGYTAHLPAP-------------------------------------
TH_4361|M.thermoresistible__bu IEATGYTASLPAPQ-------------------------------RDTGG
MAV_5246|M.avium_104 IENAGYAAAVATP-------------------------------------
MAB_3984c|M.abscessus_ATCC_199 VESVGYTAQVVGGS-------------------------------HQDAH
MLBr_01987|M.leprae_Br4923 VRRAALCT------------------------------------------
: .
Mb0994|M.bovis_AF2122/97 GGRPRGPVRGSASWPGEQNRRERRTWLPRVWLALPLGLL-ALGSSMFFGA
Rv0969|M.tuberculosis_H37Rv GGRPRGPVRGSASWPGEQNRRERRTWLPRVWLALPLGLL-ALGSSMFFGA
MMAR_1431|M.marinum_M GSCGQGAVAACHSSPGQDSGGERRKWLRRVWLTWPLALL-ATASTMVFGA
MUL_0423|M.ulcerans_Agy99 IGYSPEQDTATQLSPSEQARSGIRFWSRRAWLAWPLGLL-ASGLTMFFGA
Mflv_2244|M.gilvum_PYR-GCK --PNPPD-----VEPADPTAALRRRLIICVALSVPVIAM-AMAPPLQFTY
Mvan_4449|M.vanbaalenii_PYR-1 --PDEPA-----EPTDDPTAALKRRLLICVALSVPVIAM-AMVPVLQFTY
MSMEG_5014|M.smegmatis_MC2_155 --PQPETGGTEQDPEDRATDALRTRLLVSAVLTVPVIAM-AMVPALQFTN
TH_4361|M.thermoresistible__bu A-PAGAATGAAGDDHDAEAERWRRRLTISAALSIPVVVL-SMIPALQFTH
MAV_5246|M.avium_104 ---SPPR-------DDPELASLRRRLVTAIALAGPVIAV-AMIPALQFQH
MAB_3984c|M.abscessus_ATCC_199 AGDHDGHDPHDHMNHDVPDDQLKSRLIGSLILAIPVVVI-SMAMPLHFSG
MLBr_01987|M.leprae_Br4923 ---DGGEALQRRQADADNARYLLIRLAVAAALFVPLAHLSVMFAVLPSTH
* *: : :
Mb0994|M.bovis_AF2122/97 YPWAGWLAFAATLPVQFVAGWPILRGAVQQARALTSNMDTLIALGTLTAF
Rv0969|M.tuberculosis_H37Rv YPWAGWLAFAATLPVQFVAGWPILRGAVQQARALTSNMDTLIALGTLTAF
MMAR_1431|M.marinum_M APWAGWLAFAATVPVQFVAGWPILKAAAQQARALSANMDTLIALGTLTAF
MUL_0423|M.ulcerans_Agy99 YPWAGWLAFAATIPVQFIAGWPFLVGAVQQARARSANMNTLISLGTLTAF
Mflv_2244|M.gilvum_PYR-GCK W---QWLSLTLAAPVVVWGAWPFHRATWANLRHGTATMDTLISVGTLAAF
Mvan_4449|M.vanbaalenii_PYR-1 W---QWLSLTLAAPVVVWGAWPFHQAAWTNLRHGTATMDTLISIGTLAAL
MSMEG_5014|M.smegmatis_MC2_155 W---QWLSLTLAAPVVVWGALPFHRAAWLNLRHGTATMDTLISMGTLAAF
TH_4361|M.thermoresistible__bu W---QWLALTLTSPVVVWGAWPFHRAAVANLRHGTATMDTLISLGVTAAY
MAV_5246|M.avium_104 W---HWAALALTAPVVGWCGRPFHAAAWANLKHGVTTMDTLISIGTLAAF
MAB_3984c|M.abscessus_ATCC_199 W---EILVAALTTPILLWGGYPFHRAALVNARHGASTMDTLVSLGTIAAY
MLBr_01987|M.leprae_Br4923 FPGWEWMLTALAIPVVTWAAWPFHRVAIHNARYHGASMETLISTGITAAT
: : *: . *: : : : :.*:**:: * :*
Mb0994|M.bovis_AF2122/97 VYSTYQLFAGGP---------------------------LFFDTSALIIA
Rv0969|M.tuberculosis_H37Rv VYSTYQLFAGGP---------------------------LFFDTSALIIA
MMAR_1431|M.marinum_M GYSTYQLFAGGP---------------------------LFFDTAALIIA
MUL_0423|M.ulcerans_Agy99 IYSTYQLFAGGP---------------------------LFFDTAALIIA
Mflv_2244|M.gilvum_PYR-GCK GWSIYALFWGTAGVPGMTHPFELTI------ARTDGSGNIYLEAAAGVTT
Mvan_4449|M.vanbaalenii_PYR-1 GWSIYALFWGTAGIPGMTHPFELTI------ARTDGTSNIYLEAAAGVTT
MSMEG_5014|M.smegmatis_MC2_155 GWSLYALFWGTAGVPGMTHPFSLVG------SRTDGAGNIYLEVAAGVTT
TH_4361|M.thermoresistible__bu LWSLWALFFTAAGMPGMTMTFTLLP------ADT-GEPHIYLEVAAAVTT
MAV_5246|M.avium_104 LWSLYALVLGAADRPGMRHDFELTVGHGAHVSHVAAPCHVYFEVAAGVTL
MAB_3984c|M.abscessus_ATCC_199 LWSLWVLFANAG--------------------HTHGDVHVYFEVVAAVTV
MLBr_01987|M.leprae_Br4923 IWSLYTVFGHHQSTEHRGVWRALLG-----------SDAIYFEVAAGITV
:* : :. ::::. * :
Mb0994|M.bovis_AF2122/97 FVVLGRHLEARATGKASEAISKLLELGAKEATLLV-DGQELLVPVDQVQV
Rv0969|M.tuberculosis_H37Rv FVVLGRHLEARATGKASEAISKLLELGAKEATLLV-DGQELLVPVDQVQV
MMAR_1431|M.marinum_M FVVLGRYFEARATGKTSEAISKLLEMGAKEATLLV-DDQELLVPVDQVRV
MUL_0423|M.ulcerans_Agy99 FVVLGRYFEATAHGKTREAISKLLDMGAKETRLIV-DGEEHLVPVDRVQV
Mflv_2244|M.gilvum_PYR-GCK FILAGRYFEARAKRRAGAALRALLELGVRDVTVRR-GAAEHRIPIDQLRV
Mvan_4449|M.vanbaalenii_PYR-1 FILAGRYFEARAKRRAGAALRALLELGAREVTVRR-GGAEQRIPIEQLVI
MSMEG_5014|M.smegmatis_MC2_155 FILAGRYFEAKSKRRASAALRALMELGAKDVAILR-NGAEQRIPIDQLGV
TH_4361|M.thermoresistible__bu FIIAGRYFEARAKRQSGAALRALLDMGANDVAVLR-DGAEKRVPIGELHV
MAV_5246|M.avium_104 FVLAGRYFERRSKRTAGAALRALLALGAKDVAVLR-AGAETRIPIERLAV
MAB_3984c|M.abscessus_ATCC_199 FLLAGRFAESRAKRSAGSALRALLSLGAKDVAVLR-GGHETRIPVDQLAV
MLBr_01987|M.leprae_Br4923 FVLAGKYYTARAKSHASIALLALAALSAKDAAVLQPDGSEMVIPANELNE
*:: *:. : : *: * :...:. : * :* .:
Mb0994|M.bovis_AF2122/97 GDLVRVRPGEKIPVDGEVTDGRAAVDESMLTGESVPVEKTAGDRVAGATV
Rv0969|M.tuberculosis_H37Rv GDLVRVRPGEKIPVDGEVTDGRAAVDESMLTGESVPVEKTAGDRVAGATV
MMAR_1431|M.marinum_M GDLVRVRPGEKIPVDGEVTDGRAAVDESMLTGESVPVEKTVGDRVAGATV
MUL_0423|M.ulcerans_Agy99 GDLVRVRPGEKIPVDGEVIDGRAAVDESMLTGESVPVEKGVGDWVAGATV
Mflv_2244|M.gilvum_PYR-GCK DDEFVVRPGEKVAADGIIVEGASAVDASMLTGESVPLEVKPGDRVVGATV
Mvan_4449|M.vanbaalenii_PYR-1 GDEFVVRPGEKIAADGTVVEGASAVDASMLTGESVPEEVQPGDQVVGATV
MSMEG_5014|M.smegmatis_MC2_155 GDEFVVRPGEKIATDGVVVDGSSAVDASMLTGESVPVEVGPGDQVVGATV
TH_4361|M.thermoresistible__bu GDVFVVRPGEKIATDGEVVAGNSAVDASMLTGESVPVEVGPGDQVVGATV
MAV_5246|M.avium_104 GDEFVVRPGERIATDGIVVAGSSAVDAAMLTGESVPVEVGVGDGVTGGTV
MAB_3984c|M.abscessus_ATCC_199 GDEFVVRPGERVATDGEVIEGHSALDTSTMTGESVPMEVTVGGPVLGGYV
MLBr_01987|M.leprae_Br4923 QQRFVVRPGQTIAADGLVIDGSATVSMSPITGEAKPVRVNPGAQVIGGTV
: . ****: :..** : * :::. : :***: * . * * *. *
Mb0994|M.bovis_AF2122/97 NLDGLLTVRATAVGADTALAQIVRLVEQAQGDKAPVQRLADRVSAVFVPA
Rv0969|M.tuberculosis_H37Rv NLDGLLTVRATAVGADTALAQIVRLVEQAQGDKAPVQRLADRVSAVFVPA
MMAR_1431|M.marinum_M NTDGLLTVRATAVGADTALAQIVRLVEQAQGGKAPVQRLADRVSAVFVPV
MUL_0423|M.ulcerans_Agy99 NTDGLLTVRATAVGADTALAQIVRLVEEAQGGKAPVQRLADRVSAVFVPV
Mflv_2244|M.gilvum_PYR-GCK NVDGRLVVRAERVGADTQLAQMARLVEDAQNGKAQAQRLADRISGVFVPV
Mvan_4449|M.vanbaalenii_PYR-1 NVDGRLVVRADRVGSDTQLAQMARMVEDAQNGKAQAQRLADRISGVFVPI
MSMEG_5014|M.smegmatis_MC2_155 NVGGRLVVRANRIGSDTQLAQMARLVEDAQSGKAQAQRLADRVSAVFVPI
TH_4361|M.thermoresistible__bu NVGGHLQVRATRVGADTQLAQMARLVTEAQSGKAEVQRLADRVSAVFVPI
MAV_5246|M.avium_104 NAGGRLVVRATGIGDDTQLARMAQLVERAQSGKADAQRLADRVSGVFVPV
MAB_3984c|M.abscessus_ATCC_199 NTHGRIVVRANKVGADTQLARMARLVSDAQTGKAQVQRLADRVSSVFVPV
MLBr_01987|M.leprae_Br4923 VLNGRLIVEAAAVGDETQLAGMVRLVEQAQQQNANAQRLADRIASVFVPC
* : *.* :* :* ** :.::* ** :* .******::.****
Mb0994|M.bovis_AF2122/97 VIGVAVATFA-GWTLIAANPVAGMTAAVAVLIIACPCALGLATPTAIMVG
Rv0969|M.tuberculosis_H37Rv VIGVAVATFA-GWTLIAANPVAGMTAAVAVLIIACPCALGLATPTAIMVG
MMAR_1431|M.marinum_M VVGVAAATFV-GWSLIAADPVGGMTAAVAVLIIACPCALGLATPTAITVG
MUL_0423|M.ulcerans_Agy99 VVRIAAATFA-GWTLLAANPVAGMTAAVSVLIIACPYALGLATPTAIIVG
Mflv_2244|M.gilvum_PYR-GCK VLGLAVATLG-FWIGSGYPVAAAFTAAVAVLIIACPCALGLATPMALMVG
Mvan_4449|M.vanbaalenii_PYR-1 VIALSVATLG-FWIGSGFPVAAAFTAAVAVLIIACPCALGLATPTALMVG
MSMEG_5014|M.smegmatis_MC2_155 VIALAVATLG-FWLGTGGSVAAAFTAAVAVLIIACPCALGLATPTALLVG
TH_4361|M.thermoresistible__bu VIALSALTLAGWLLLTDAPVSAAFTAAVAVLIIACPCALGLATPTALLVG
MAV_5246|M.avium_104 VLLLAVATLA-GWLAAGGTLATALTAAVAVLIIACPCALGLATPTALLVG
MAB_3984c|M.abscessus_ATCC_199 VLAISALTFV-AWAALGNSLASAFTAAVAVLIIACPCALGLATPTAILVG
MLBr_01987|M.leprae_Br4923 VFAVAALTAV-GWLIAGSGPDRVFSAAIAVLVIACPCALGLATPTAMMVA
*. ::. * ::**::**:**** ******* *: *.
Mb0994|M.bovis_AF2122/97 TGRGAELGILVKGGEVLEASKKIDTVVFDKTGTLTRARMRVTDVIAGQRR
Rv0969|M.tuberculosis_H37Rv TGRGAELGILVKGGEVLEASKKIDTVVFDKTGTLTRARMRVTDVIAGQRR
MMAR_1431|M.marinum_M TGRGAEMGILVKGGEVLEASKKIDTVVFDKTGTLTRAQMRLTDVIAGKRR
MUL_0423|M.ulcerans_Agy99 TGRGADMGILVKGGEVLEASKKIDTVVFDKTGTLTRAQMELTDVIAGKRR
Mflv_2244|M.gilvum_PYR-GCK TGRGAQLGILIKGPEVLESTRRVDTVIVDKTGTVTTGDMTLVDVFAADGE
Mvan_4449|M.vanbaalenii_PYR-1 TGRGAQLGILIKGPEVLEDTRRVDTVILDKTGTVTTGSMTLVDVFAAEGE
MSMEG_5014|M.smegmatis_MC2_155 TGRGAQLGILIKGPEVLESTRRVDTIVLDKTGTITTGSMTLLDVVVADGE
TH_4361|M.thermoresistible__bu TGRGAQLGILIKGPQVLESTRRIDTVVLDKTGTVTTGQMSVTSVHVADGL
MAV_5246|M.avium_104 TGRAAQLGVLIKGPEVLETTRAVDTVVLDKTGTVTTGAMTVLDVVAADGT
MAB_3984c|M.abscessus_ATCC_199 TGRGAQLGVLIKGPEVLENVKDIDTVVLDKTGTVTTGVMSVVEVRRSIGS
MLBr_01987|M.leprae_Br4923 SGRGAQLGILLKGHESFEATRAVDTVVFDKTGTLTTGQLKVSAVTAAPGW
:**.*::*:*:** : :* : :**::.*****:* . : : * .
Mb0994|M.bovis_AF2122/97 ---QPNQVLRLAAAVESGSEHPIGAAIVAAAHERGLAIPAANAFTAVAGH
Rv0969|M.tuberculosis_H37Rv ---QPDQVLRLAAAVESGSEHPIGAAIVAAAHERGLAIPAANAFTAVAGH
MMAR_1431|M.marinum_M ---QPDPVLRIAAAVESGSEHPIGAAIVAGARERGLEIPSATGFANLAGH
MUL_0423|M.ulcerans_Agy99 ---QPDVVLRIAAAVESGSEHPIGAAIVAGARERELEVPPATEFTNLAGH
Mflv_2244|M.gilvum_PYR-GCK ---QPDEVLRMAGAVESPSEHPIAAAIAGGARDKLGDLPAVDGFANLRGL
Mvan_4449|M.vanbaalenii_PYR-1 ---QPDEVLRVAGAVESSSEHPIARAIAAGAQEKFGELPAVTAFTNLRGL
MSMEG_5014|M.smegmatis_MC2_155 ---DRDDVLRVAAAVEDASEHPIARAIAAGARERTGELPAVEGFANIEGL
TH_4361|M.thermoresistible__bu ---SETELLRLAGAVEHASEHPIARAVATHAAQTVPPLPEVSDFENHGGN
MAV_5246|M.avium_104 ---DRATLLRYAGALEAASEHPIAHAIARDAKAELGPLPTPTGFRAVGGG
MAB_3984c|M.abscessus_ATCC_199 GVADADEILRIAASVESASEHPVARAIVAAAASARLTLSPVTNFRNEPGV
MLBr_01987|M.leprae_Br4923 ---QANEVLQMAATVESASEHAVALAIAASTTHR----EPVANFRAVPGH
. :*: *.::* ***.:. *:. : * *
Mb0994|M.bovis_AF2122/97 GVRAQVNGGPVVVGRRKLVDEQHLVLPDHLAAAAVEQEERGRTAVFVGQD
Rv0969|M.tuberculosis_H37Rv GVRAQVNGGPVVVGRRKLVDEQHLVLPDHLAAAAVEQEERGRTAVFVGQD
MMAR_1431|M.marinum_M GVRAEVNGKPVLVGRRKLVDESDLVLPDHLAAAAAELEQQGRTAVFVGQD
MUL_0423|M.ulcerans_Agy99 GVRGEVGGKSVLVGRRKLVDEQHLRLPEQLAAAAADLEERGRTAVYVGQD
Mflv_2244|M.gilvum_PYR-GCK GVEGDVEGRRLTLGRLRLLSDRSFEIPDDLASAVARAEAGGSTAVTVGWD
Mvan_4449|M.vanbaalenii_PYR-1 GVSGQVDGRAVLLGRLRLLADRSHEIPSELEVAVARAESDGRTAVVVGWD
MSMEG_5014|M.smegmatis_MC2_155 GVQGTVEGRAVVVGRPRLFGDPEKSLPRKLADAMEAAQAQGRTAVVVGWD
TH_4361|M.thermoresistible__bu GVSGRVDGHDVRVGRLAWLG---LSAGDRLRNAAAQAEAAGHTPVWVAVD
MAV_5246|M.avium_104 GVHGRVDGHAVAIGRPSWLAERGLRPDAALAAAAARAEHDGKTVVAVGWD
MAB_3984c|M.abscessus_ATCC_199 GVSGVVDGQEVSI----------------------EAIEGQNTTAAVSWG
MLBr_01987|M.leprae_Br4923 GVSGTVAERAVRVGKPSWIASR--CNSTTLVTARRNAELRGETAVFVEID
** . * : : * . * .
Mb0994|M.bovis_AF2122/97 GQVVGVLAVADTVKDDAADVVGRLHAMGLQVAMITGDNARTAAAIAKQVG
Rv0969|M.tuberculosis_H37Rv GQVVGVLAVADTVKDDAADVVGRLHAMGLQVAMITGDNARTAAAIAKQVG
MMAR_1431|M.marinum_M DQVVGVLAVADTVKDDAIEVVRRLHAIGLKVAMITGDNARTAAAIAQQVG
MUL_0423|M.ulcerans_Agy99 GQVVGVLAVADTVKDDAADVVSQLHAMGLQVALITGDNARTAAAIAAQVG
Mflv_2244|M.gilvum_PYR-GCK GRARGVLMVADAVKPTSAEAISLLKRAGLTPIMVTGDNEAVARTVAAQVG
Mvan_4449|M.vanbaalenii_PYR-1 GQARGVLMVADAVKPTSAEAISLLKRLGLTPIMVTGDNEAVARAVAAEVG
MSMEG_5014|M.smegmatis_MC2_155 GTARGVLVVADAVKATSAEAIGQLRKLGLQPIMLTGDNGAAARAIADQVG
TH_4361|M.thermoresistible__bu GRIAGVIVVSDTVKDHAAEAIAELRALGATPVLLTGDNDRAARTVAAQVG
MAV_5246|M.avium_104 GRARGILALADTVKPSSAAAVRQFTRLGLTPILLTGDNHTVARRTAGELG
MAB_3984c|M.abscessus_ATCC_199 GLRRGEIEVADTVKPSSRRAIDELKAMGITPVLLTGDNAEVAARVASEVG
MLBr_01987|M.leprae_Br4923 GEQCGVIAVADAVKASAADAVAALHDRGFRTALLTGDNPASAAAVASRIG
. * : ::*:** : .: : * ::**** * * .:*
Mb0994|M.bovis_AF2122/97 IE--KVLAEVLPQDKVAEVRRLQDQGRVVAMVGDGVNDAPALVQADLGIA
Rv0969|M.tuberculosis_H37Rv IE--KVLAEVLPQDKVAEVRRLQDQGRVVAMVGDGVNDAPALVQADLGIA
MMAR_1431|M.marinum_M ID--QVLAEVLPADKVTEIRRLQNQGQVVAMVGDGVNDAPALVQADLGIA
MUL_0423|M.ulcerans_Agy99 ID--RVLAEVLPADKVNEVRRLQDEGRIVAMVGDGVNDAPALVQADLGIA
Mflv_2244|M.gilvum_PYR-GCK ID--EVVAGVLPQDKVDTVTRLQRDGRVVAMVGDGVNDAAALAQADLGMA
Mvan_4449|M.vanbaalenii_PYR-1 ID--EVIAGVLPQDKVDTVKRLQRDGKVVAMVGDGVNDAAALAQADLGLA
MSMEG_5014|M.smegmatis_MC2_155 ID--DVYAEVLPQDKADVIKRLQAEGRVVAMVGDGVNDAAALAQADLGLS
TH_4361|M.thermoresistible__bu ID--DVIAGVLPAEKVDHIKQLQDRGRVVAMIGDGVNDAAALAQADLGLA
MAV_5246|M.avium_104 IG--EVISGALPADKVEAVKRLQSAGRVVAMVGDGVNDAAALAAADLGLA
MAB_3984c|M.abscessus_ATCC_199 IAAGDVIAGVKPTEKADVVKRLQAEGKRVAMVGDGVNDSAALATADIGMA
MLBr_01987|M.leprae_Br4923 ID--EVIADILPEDKVDVIEQLRDRGHVVAMVGDGINDGPALARADLGMA
* * : * :*. : :*: *: ***:***:**..**. **:*::
Mb0994|M.bovis_AF2122/97 IGTGTDVAIEASDITLMSGRLDGVVRAIELSRQTLRTIYQNLGWAFGYNT
Rv0969|M.tuberculosis_H37Rv IGTGTDVAIEASDITLMSGRLDGVVRAIELSRQTLRTIYQNLGWAFGYNT
MMAR_1431|M.marinum_M IGTGTDVAIEASDITLMSGQLDGVVRAIGLSRQTLRTIYQNLGWAFGYNT
MUL_0423|M.ulcerans_Agy99 IGTGTDVAIEASDITLMSGRLDGVVSAIELSRRTLRTIYQNLGWAFGYNT
Mflv_2244|M.gilvum_PYR-GCK MGSGTDVAIEASDLTLVSNDLRAVPDAIRLSRKTLSTIKSNLFWAFAYNV
Mvan_4449|M.vanbaalenii_PYR-1 MGSGTDVAIEASDLTLVSNDLRAVPDAIRLSRKTLSTIKGNLFWAFAYNV
MSMEG_5014|M.smegmatis_MC2_155 MGTGTDVAIEAGDLTLVRGDLRTAADAIRLSRRTLATIKGNLFWAFAYNV
TH_4361|M.thermoresistible__bu IGTGTDVAIEASDLTLVTGDLRAAPDAIRLGRATLRTIKGNLFWAFAYNV
MAV_5246|M.avium_104 MGTGTDVAIEAADVTVVRGDLRAAVDAIRLSRRTLATIKTNLVWAFGYNL
MAB_3984c|M.abscessus_ATCC_199 MGTGTDAAIEASDLTLVRGDLTTVPTALRLSRKTLGTIKANLFWAFAYNT
MLBr_01987|M.leprae_Br4923 IGRGTDVAIGAADIILVRDNLDVVPITLDLAAATMRTIKFNMVWAFGYNI
:* ***.** *.*: :: . * . :: *. *: ** *: ***.**
Mb0994|M.bovis_AF2122/97 AAIPLAALGALNPVVAGAAMGFSSVSVVTNSLRLRRFGRDGRTA------
Rv0969|M.tuberculosis_H37Rv AAIPLAALGALNPVVAGAAMGFSSVSVVTNSLRLRRFGRDGRTA------
MMAR_1431|M.marinum_M AAIPLAALGLLNPVIAGAAMGLSSVSVVTNSLRLRRFGRDAQQQPERHTA
MUL_0423|M.ulcerans_Agy99 AAIPLAALGVLNPVVAGAAMGLSSVSVVTNSLRLRRFGRDV---------
Mflv_2244|M.gilvum_PYR-GCK AALPLAAAGLLNPMIAGAAMAFSSVFVVTNSLRLRAFTPSR---------
Mvan_4449|M.vanbaalenii_PYR-1 AALPLAAAGLLNPMIAGAAMAFSSVFVVSNSLRLRAFTPSR---------
MSMEG_5014|M.smegmatis_MC2_155 AALPLAAAGLLNPMLAGAAMAFSSVFVVSNSLRLKRFR------------
TH_4361|M.thermoresistible__bu AALPLAAAGLLNPLIAGAAMAFSSVFVVSNSLRLRRFTPSR---------
MAV_5246|M.avium_104 AAIPLAALGMLNPMLAGAAMALSSVLVVGNSLRLRSFASIIPGA------
MAB_3984c|M.abscessus_ATCC_199 AAIPLAALGLLNPMIAGAAMAMSSVFVVLNSLRLRTFR------------
MLBr_01987|M.leprae_Br4923 AAIPIAAAGLLNPLVAGAAMAFSSFFVVSNSLRLRNFGAILSCGTSRHRT
**:*:** * ***::*****.:**. ** *****: *
Mb0994|M.bovis_AF2122/97 ----------------------------------------
Rv0969|M.tuberculosis_H37Rv ----------------------------------------
MMAR_1431|M.marinum_M VAPIRT----------------------------------
MUL_0423|M.ulcerans_Agy99 ----------------------------------------
Mflv_2244|M.gilvum_PYR-GCK ----------------------------------------
Mvan_4449|M.vanbaalenii_PYR-1 ----------------------------------------
MSMEG_5014|M.smegmatis_MC2_155 ----------------------------------------
TH_4361|M.thermoresistible__bu ----------------------------------------
MAV_5246|M.avium_104 ----------------------------------------
MAB_3984c|M.abscessus_ATCC_199 ----------------------------------------
MLBr_01987|M.leprae_Br4923 VKRWRCPPPTRLRSTACSPVDASPLRPVAHRTGVKPPTHR