For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AFLFVAPETLTAAAADIAGIGSTLGEAGAAAATPTTAVLAAGADDVSAAVATLFSSYGQAYQSLSAQAAQ FHSQFVQAMQAGAGSYATAEASNVAQTALDLINQPVLTLTGRPLIGNGANGAAGTGANGGAAGWLLGDGG AGGSGGLEQQGGTGGAAGLFGNGGAGGAGGASTVGTGAAGGAGGAGGLLWGQGGIGGTGGSGIDGGAGGA GGAGGALFGIGGAGGQGGIASTGLTGGGAGGDGGAGGLFGNGGIGGTGGLASVGPGGVGGNGGAAGTLLG NGGGGGVGGFGATQGGDGGDGGATGIFGGTGGAGGAGGQSTNALADSVGGNGGQGGDGRLFGNGGAGGVG GAAYTSNILMNATGGNGGQGGTGGTLFGNGGGGGTGGAGFVGPSSAGDGGNGGGGGRAGLIGNGGAGGAG GAPGPNGGFSGGNGGNGGDAVLIGNGGNSGDVGLSGAGTPGLPGNGGLLIGTIGNT*
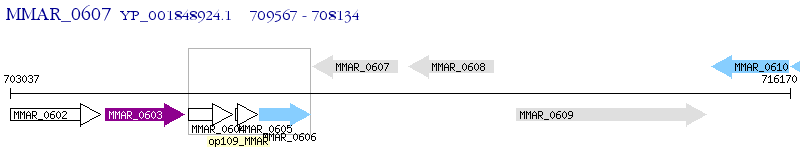
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0607 | - | - | 100% (477) | PE-PGRS family protein, PE_PGRS9_5 |
| M. marinum M | MMAR_4398 | - | e-143 | 57.52% (499) | PE-PGRS family protein, PE_PGRS9_3 |
| M. marinum M | MMAR_4399 | - | e-142 | 57.09% (494) | PE-PGRS family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1849c | PE_PGRS33 | 1e-144 | 57.45% (510) | PE-PGRS family protein |
| M. gilvum PYR-GCK | Mflv_5091 | - | 5e-86 | 45.47% (486) | triple helix repeat-containing collagen |
| M. tuberculosis H37Rv | Rv1818c | PE_PGRS33 | 1e-143 | 57.37% (509) | PE-PGRS family protein |
| M. leprae Br4923 | MLBr_01183 | - | 2e-12 | 36.96% (92) | hypothetical protein MLBr_01183 |
| M. abscessus ATCC 19977 | MAB_4284c | - | 7e-41 | 36.59% (369) | hypothetical protein MAB_4284c |
| M. avium 104 | MAV_1346 | - | 5e-22 | 35.21% (267) | PE family protein |
| M. smegmatis MC2 155 | MSMEG_0868 | - | 2e-66 | 42.58% (418) | hypothetical protein MSMEG_0868 |
| M. thermoresistible (build 8) | TH_3810 | - | 2e-39 | 39.67% (300) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_0212 | - | 1e-135 | 51.85% (540) | PE-PGRS family protein |
| M. vanbaalenii PYR-1 | Mvan_1264 | - | 4e-81 | 43.71% (485) | triple helix repeat-containing collagen |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1849c|M.bovis_AF2122/97 --------------------------------------------------
Rv1818c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_0212|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0607|M.marinum_M --------------------------------------------------
Mflv_5091|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1264|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0868|M.smegmatis_MC2_155 MHERLRERAGGVRKTLSDFAEGAGLKPSDTDDTRSPLRKPKFPAALAPES
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01183|M.leprae_Br4923 --------------------------------------------------
MAV_1346|M.avium_104 --------------------------------------------------
Mb1849c|M.bovis_AF2122/97 --------------------------------------------------
Rv1818c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_0212|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0607|M.marinum_M --------------------------------------------------
Mflv_5091|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1264|M.vanbaalenii_PYR-1 -------------------MMFLAPPGGPGPISAAIQLTSTGSEATEP--
MSMEG_0868|M.smegmatis_MC2_155 PLPDLKPRWERPTTAPDSETPSITQPQGRDTAADPAEQQATSTNTTDQPT
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01183|M.leprae_Br4923 --------------------------------------------------
MAV_1346|M.avium_104 --------------------------------------------------
Mb1849c|M.bovis_AF2122/97 ---------------------------------------MSFVVTIP---
Rv1818c|M.tuberculosis_H37Rv ---------------------------------------MSFVVTIP---
MUL_0212|M.ulcerans_Agy99 ---------------------------------------MSFVNVTP---
MMAR_0607|M.marinum_M ---------------------------------------MAFLFVAP---
Mflv_5091|M.gilvum_PYR-GCK --------------------------------------MVTFGGGATPTP
Mvan_1264|M.vanbaalenii_PYR-1 ------------------------------------WYVVSFGGSGAPTP
MSMEG_0868|M.smegmatis_MC2_155 SDSLGAQPTAQVARQVQAPATDPAPAAPSLRPISGLLSVVTLGTTLIPNA
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 ------------------------------------MSIIDFILNLFRDP
MLBr_01183|M.leprae_Br4923 --------------------------------------MPLFLNAEP---
MAV_1346|M.avium_104 --------------------------------------MS-FVTTRP---
Mb1849c|M.bovis_AF2122/97 ----------------------------------------EALAAVATDL
Rv1818c|M.tuberculosis_H37Rv ----------------------------------------EALAAVATDL
MUL_0212|M.ulcerans_Agy99 ----------------------------------------EAVAAAASDL
MMAR_0607|M.marinum_M ----------------------------------------ETLTAAAADI
Mflv_5091|M.gilvum_PYR-GCK ----------------------------------------TSLGAAAVDL
Mvan_1264|M.vanbaalenii_PYR-1 ----------------------------------------NNLAASLVDL
MSMEG_0868|M.smegmatis_MC2_155 PTEPADMPGVWAVMAWARRQPMYSDPAMRTTVNPQAQLPVPTVASAVLDP
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 ----------------------------------------VEAASYVADP
MLBr_01183|M.leprae_Br4923 ----------------------------------------QALTAAANTL
MAV_1346|M.avium_104 ----------------------------------------EALLAVASML
Mb1849c|M.bovis_AF2122/97 AG------------------------------------------------
Rv1818c|M.tuberculosis_H37Rv AG------------------------------------------------
MUL_0212|M.ulcerans_Agy99 TG------------------------------------------------
MMAR_0607|M.marinum_M AG------------------------------------------------
Mflv_5091|M.gilvum_PYR-GCK S------------------------GTSCG----------LICNGQDGTG
Mvan_1264|M.vanbaalenii_PYR-1 S------------------------GTSCG----------LICNGQDGTG
MSMEG_0868|M.smegmatis_MC2_155 TQIVNWLIYQPLHAAGQQFWVVSPIGSTLGGVINTISGQYLIGNGINGTA
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 EAA-------------------------------------LANAGLSAVS
MLBr_01183|M.leprae_Br4923 EG------------------------------------------------
MAV_1346|M.avium_104 EG------------------------------------------------
Mb1849c|M.bovis_AF2122/97 ------IGSTIGTANAAAAVPTTTVLAAAADEVSAAMAALFSGHAQAYQA
Rv1818c|M.tuberculosis_H37Rv ------IGSTIGTANAAAAVPTTTVLAAAADEVSAAMAALFSGHAQAYQA
MUL_0212|M.ulcerans_Agy99 ------IGSVLGEANAVAARATTTVLAAGGDEVSAAIAALFSFHGQSYQT
MMAR_0607|M.marinum_M ------IGSTLGEAGAAAATPTTAVLAAGADDVSAAVATLFSSYGQAYQS
Mflv_5091|M.gilvum_PYR-GCK QN-----GGILFGNGGSGWNSDVVGMDGGNGGNGGLFGGNGGNGGNGLGG
Mvan_1264|M.vanbaalenii_PYR-1 QN-----GGILFGNGGSGWNSEIAGVSGGNGGNAGLFGGNGGNGGNGLGS
MSMEG_0868|M.smegmatis_MC2_155 DNPNGGTGGLLFGDGGHGYDGSAAGLAGGNGGNAGLFG-NGGNGGVGWAG
TH_3810|M.thermoresistible__bu ------------------------------------------------VD
MAB_4284c|M.abscessus_ATCC_199 AAQVGAVMPVVAESVASNYGYQSQWVNTGN----PIHDLSGNLQNYYAAQ
MLBr_01183|M.leprae_Br4923 ------LSAATVASNAAAAQLTTEIAPPAADDVSILLAHFFSGHGRQYQA
MAV_1346|M.avium_104 ------LGSSMDAQNAAAAAPTTSIAPAAADEVSALQAAQFSAYGTWYQQ
Mb1849c|M.bovis_AF2122/97 LSAQAALFHEQFVRALTAGAGSYAAAEAASAAPLEGVLDVINAPALALLG
Rv1818c|M.tuberculosis_H37Rv LSAQAALFHEQFVRALTAGAGSYAAAEAASAAPLEGVLDVINAPALALLG
MUL_0212|M.ulcerans_Agy99 LGAQAEAFHAQFAQLMANGAGTYASAEAANAAPFQGLLGAVNAPVQTLTG
MMAR_0607|M.marinum_M LSAQAAQFHSQFVQAMQAGAGSYATAEASNVA--QTALDLINQPVLTLTG
Mflv_5091|M.gilvum_PYR-GCK GNGGNGGNAGAFALFGNGGNGGNGGVGVVGATGAVG--APGVAGATGATG
Mvan_1264|M.vanbaalenii_PYR-1 GDGGNGGDAGLFSLFGRGGNGGNGGVGIAGATGAVG--APGVAGATGATG
MSMEG_0868|M.smegmatis_MC2_155 LDGGNGGSGG--LLMGIGGAGGAGGVGADGTSAHVDGGTGGTGGTGGGTG
TH_3810|M.thermoresistible__bu GDGGSGGRTGLFSKLSRGGDGGDGGLG-------------GNGGHGGGTG
MAB_4284c|M.abscessus_ATCC_199 PDGPSILSPSLLSPRDNDFLSHNDTDFASHNPIASGNQVMSPSGNVDSFN
MLBr_01183|M.leprae_Br4923 HASQGATNHQDLIQSLLTSSSAYAGTETAN--------------------
MAV_1346|M.avium_104 VSAQAKAIHQTLVQNLDTNADSYGATEAANRVNTSAAALQGLAPNVIPNA
. .
Mb1849c|M.bovis_AF2122/97 RPLIGN----------------GANGAPGTGANGG--------------D
Rv1818c|M.tuberculosis_H37Rv RPLIGN----------------GANGAPGTGANGG--------------D
MUL_0212|M.ulcerans_Agy99 RPLIGN----------------GANGAPGTGANGG--------------A
MMAR_0607|M.marinum_M RPLIGN----------------GANGAAGTGANGG--------------A
Mflv_5091|M.gilvum_PYR-GCK VTSGVVGGNGEDAVGYGATGAAGAAGAAGAAGAAG--------------A
Mvan_1264|M.vanbaalenii_PYR-1 VTSNVTGGNGADAVGYGATGAAGAAGAAGAAGKAG--------------A
MSMEG_0868|M.smegmatis_MC2_155 GTGGIGGTGGGLFGLLGHSGSTGAAGAAGNGGDGGNGGFGYVLIAGPNVT
TH_3810|M.thermoresistible__bu IIS--------------------LLSLAGNGGNGG---------------
MAB_4284c|M.abscessus_ATCC_199 RGPLLDLSFGDLQFGNRDTTAIGDGAVAVGGSNSGP----------IASG
MLBr_01183|M.leprae_Br4923 --------------------------HDSL--------------------
MAV_1346|M.avium_104 APAADP----------------PPAGTTSLGTS-----------------
Mb1849c|M.bovis_AF2122/97 GGILIGNGGAGGSGAAG---------------------------------
Rv1818c|M.tuberculosis_H37Rv GGILIGNGGAGGSGAAG---------------------------------
MUL_0212|M.ulcerans_Agy99 GGWLIGNGGAGGSGAPG---------------------------------
MMAR_0607|M.marinum_M AGWLLGDGGAGGSGGLE---------------------------------
Mflv_5091|M.gilvum_PYR-GCK VGSAGAAGEAGETGTWG--NAGLPGKAGDAGGAG----------------
Mvan_1264|M.vanbaalenii_PYR-1 VGSAGTAGSAGAPGVWG--NAGLPGAAGAAGGAG----------------
MSMEG_0868|M.smegmatis_MC2_155 VGSETRGGTGGAGGTGGQSTAAGPGKGGGGGGGGGGAILIGANGGTVDGH
TH_3810|M.thermoresistible__bu NGYRTTDNNALDGGHGG---------------------------------
MAB_4284c|M.abscessus_ATCC_199 QGSTAVNGDVRDSNIIN---------------------------------
MLBr_01183|M.leprae_Br4923 --------------------------------------------------
MAV_1346|M.avium_104 IGWAQNVGAAASDFIT----------------------------------
Mb1849c|M.bovis_AF2122/97 -----------------------------------------MPGGNGGAA
Rv1818c|M.tuberculosis_H37Rv -----------------------------------------MPGGNGGAA
MUL_0212|M.ulcerans_Agy99 -----------------------------------------LNGGAGGAA
MMAR_0607|M.marinum_M -----------------------------------------QQGGTGGAA
Mflv_5091|M.gilvum_PYR-GCK AVGAAGAAG------TVGTAGATGGTG---------------TGANGGAG
Mvan_1264|M.vanbaalenii_PYR-1 AIGTAGVAG------AAGTAGATGGTG---------------VGGNGGAG
MSMEG_0868|M.smegmatis_MC2_155 AIGGNGGNGGNGSTTAAGGNGGTGGTGRVIAEGTGSTAEGTATGGDGGHS
TH_3810|M.thermoresistible__bu ------------------------------------------NGGNVGFF
MAB_4284c|M.abscessus_ATCC_199 ----------------------------------------ADHGSNVGVD
MLBr_01183|M.leprae_Br4923 --------------------------------------------------
MAV_1346|M.avium_104 -------------------------------------------LGEGQFA
Mb1849c|M.bovis_AF2122/97 GLFGNGG--------------------AGGAGGNVASGTAGFGGAGGAG-
Rv1818c|M.tuberculosis_H37Rv GLFGNGG--------------------AGGAGGNVASGTAGFGGAGGAG-
MUL_0212|M.ulcerans_Agy99 GLIGTGG--------------------AGGAGGSSSTVNGGLGGTGGAG-
MMAR_0607|M.marinum_M GLFGNGG--------------------AGGAGGASTVGTGAAGGAGGAG-
Mflv_5091|M.gilvum_PYR-GCK GVGGVGGTGAAG--------------AAGGAGGLGETGTTGGAGGVGGQ-
Mvan_1264|M.vanbaalenii_PYR-1 QAGGIGGTGAAG--------------AAGQAGTAGETGVTGGVGGIGGQ-
MSMEG_0868|M.smegmatis_MC2_155 TGTGIGGNGGIGSVYAYGNNGAVSGTATGGRGGDGIDGGTGGAGGMGEIY
TH_3810|M.thermoresistible__bu SLFNLGG--------------------AGGWGGSGVDG------------
MAB_4284c|M.abscessus_ATCC_199 QSQTRVGGDYTN---------------VSGHGSADIDKSVTVVDDHSQRN
MLBr_01183|M.leprae_Br4923 --------------------------------------------------
MAV_1346|M.avium_104 PVPGAAGIPNFN----------------PGLTAATTAGPPAPAAPAAAA-
Mb1849c|M.bovis_AF2122/97 ----GLLYGAGGAGGAG---------------------------------
Rv1818c|M.tuberculosis_H37Rv ----GLLYGAGGAGGAG---------------------------------
MUL_0212|M.ulcerans_Agy99 ----GWLLGNGGAGGAGGASAIPLGPDLIG--------------------
MMAR_0607|M.marinum_M ----GLLWGQGGIGGTG---------------------------------
Mflv_5091|M.gilvum_PYR-GCK -GTAGGTGGVGGIGGAGSTGAVGGVGGKG----IQGGQG-----------
Mvan_1264|M.vanbaalenii_PYR-1 -GTEGGTGGVGGVGGVGATGGAGGVGGKG----IQGGQG-----------
MSMEG_0868|M.smegmatis_MC2_155 SGADGEATGISAGGDGGSATGADSLGGRGGTSWVQAGYGPTTGGKASGTA
TH_3810|M.thermoresistible__bu ---------VDVVGGNP---------------------------------
MAB_4284c|M.abscessus_ATCC_199 TTNINDSFNTDRSTNIGSGNTTNTEIGSHN--------------------
MLBr_01183|M.leprae_Br4923 --------------------------------------------------
MAV_1346|M.avium_104 --------------------------------------------------
Mb1849c|M.bovis_AF2122/97 -------GRAGG---------------------------------GVGGI
Rv1818c|M.tuberculosis_H37Rv -------GRAGG---------------------------------GVGGI
MUL_0212|M.ulcerans_Agy99 ------KGGAGGPGG----------------------------LFGAGGL
MMAR_0607|M.marinum_M ---------------------------------------------GSGID
Mflv_5091|M.gilvum_PYR-GCK --LQGGKGGDGGAGGKGGDADSMGS------------------AGATGGK
Mvan_1264|M.vanbaalenii_PYR-1 --LQGGQGGDGGTGGTGGDATSMGE------------------AGATGGK
MSMEG_0868|M.smegmatis_MC2_155 KAGHGGKASDGGKGGNGGDALVMAAGATGVATGNTAIGGNGGAASANGAV
TH_3810|M.thermoresistible__bu ---------DGGTPG------------------------------AHGES
MAB_4284c|M.abscessus_ATCC_199 -------QANIGTS------------------------------TSAGGS
MLBr_01183|M.leprae_Br4923 --------------------------------------------------
MAV_1346|M.avium_104 ---------------------------------------------GAPVL
Mb1849c|M.bovis_AF2122/97 GGAGGAGGNGG---------------------------------------
Rv1818c|M.tuberculosis_H37Rv GGAGGAGGNGG---------------------------------------
MUL_0212|M.ulcerans_Agy99 GGAGGFGGSGGGAG------------------------------------
MMAR_0607|M.marinum_M GGAGGAGGAGG---------------------------------------
Mflv_5091|M.gilvum_PYR-GCK GGNGGNGGRGGLFG---------------KGGNAGNGAD-----------
Mvan_1264|M.vanbaalenii_PYR-1 GGNGGNGGRGSLFG---------------RGGNAGNGGN-----------
MSMEG_0868|M.smegmatis_MC2_155 GGNGGFTNIAAIHGSITDSSATSGHGGAAQGGNGGNGGNGWVQAYDPNGP
TH_3810|M.thermoresistible__bu HTPGVYTDRQ----------------------------------------
MAB_4284c|M.abscessus_ATCC_199 AAVGGNAGLAGNAG------------------------------------
MLBr_01183|M.leprae_Br4923 --------------------------------------------------
MAV_1346|M.avium_104 ASMGRGPSIGGLSVP-----------------------------------
Mb1849c|M.bovis_AF2122/97 -------------------------LLFGAGG------------------
Rv1818c|M.tuberculosis_H37Rv -------------------------LLFGAGG------------------
MUL_0212|M.ulcerans_Agy99 ---------------GQGGAGGLLSGLVGAGGGHGGTG------------
MMAR_0607|M.marinum_M -------------------------ALFGIGG------------------
Mflv_5091|M.gilvum_PYR-GCK -----------GGTGGAGGAGGTGG-TGGAGGTGG---------------
Mvan_1264|M.vanbaalenii_PYR-1 -----------GGTGGNGGNGGTGG-TGGNGGTGG---------------
MSMEG_0868|M.smegmatis_MC2_155 AGTTIDKGKAAGGNGGDGGFNGTGGATGGRGGDGAGASVWKANGQHSTGQ
TH_3810|M.thermoresistible__bu ---------------GEDGEDGTGTGIGGRGGNGG---------------
MAB_4284c|M.abscessus_ATCC_199 -------------LAGNAGLAGNAGLAGNAGLAGN---------------
MLBr_01183|M.leprae_Br4923 --------------------------------------------------
MAV_1346|M.avium_104 ----------------------PSWAPGGVAPAAN---------------
Mb1849c|M.bovis_AF2122/97 --AGSVGGLAADAG------DGGAGGDGG---LFFGVGGAGGAGGTGTNV
Rv1818c|M.tuberculosis_H37Rv --AGGVGGLAADAG------DGGAGGDGG---LFFGVGGAGGAGGTGTNV
MUL_0212|M.ulcerans_Agy99 GYGGGAGGAGATGGGN----TGGIGGDGGSAGMLFSNSGAGGAGGASELA
MMAR_0607|M.marinum_M --AGGQGGIASTGLT-----GGGAGGDGG-AGGLFGNGGIGGTGGLASVG
Mflv_5091|M.gilvum_PYR-GCK --TGGTGGIGGAGSTGGTGGTGGTGGTGGTGGTG-GTGGTGGTGGTGGKG
Mvan_1264|M.vanbaalenii_PYR-1 --TGGTGGIGGAGSTGGTGGTGGTGGTGGTGGTG-GTGGTGGTGGTGGKG
MSMEG_0868|M.smegmatis_MC2_155 TDTGGNGGRGGDGGTFVNAGANGNGGAGGAGGVSEGEGGTGGHGGDGGRG
TH_3810|M.thermoresistible__bu -SSSGTHPMAEDG--------EHVKALDGRPGLD---GGNGSHGGNGGHG
MAB_4284c|M.abscessus_ATCC_199 AGLAGNAGLAGNAGLAGNAGLAGNAGLAGNAGLA----GNAGLAGNAGLA
MLBr_01183|M.leprae_Br4923 --------------------------------------------------
MAV_1346|M.avium_104 ---ATPAQLVGAGFTSAAPDTAPVTAVPGGVPSMATAGRGGYGLGAPRYG
Mb1849c|M.bovis_AF2122/97 -TGGAGGAGGNGGLLFGAGGVGGVGGDGVAFLG------TAPGGPGGAGG
Rv1818c|M.tuberculosis_H37Rv -TGGAGGAGGNGGLLFGAGGVGGVGGDGVAFLG------TAPGGPGGAGG
MUL_0212|M.ulcerans_Agy99 -DGGAGGAGGNGGWLLSNGGVGGAGGAGADAVGPPFGIPAGSGSGGGAGG
MMAR_0607|M.marinum_M -PGGVGGNGGAAGTLLGNGGGGGVGGFG-----------ATQGGDGGDGG
Mflv_5091|M.gilvum_PYR-GCK GTGGTGGAGGTG----GDGGTGGTGGTGGTG---------GTGGTGGTGG
Mvan_1264|M.vanbaalenii_PYR-1 GTGGTGGDGGTG----GTGGTGGTGGTGGTG---------GTGGTGGTGG
MSMEG_0868|M.smegmatis_MC2_155 GDGALGGNGGTGGVGLGNGGKGGNGGNGGNGGTYVDPSHTGAGGNGGNGG
TH_3810|M.thermoresistible__bu GRGGFLGIGGSG-------GWGGHGGNGGD-----------TDADGGNGG
MAB_4284c|M.abscessus_ATCC_199 GNAGLAGNAGLAG----NAGLAGNAGLAGNAG--------LAGNAGLAGN
MLBr_01183|M.leprae_Br4923 --------------------------------------------------
MAV_1346|M.avium_104 VKPTVMPKPAI---------------------------------------
Mb1849c|M.bovis_AF2122/97 AGG------------LFGVGGAGG--------------------------
Rv1818c|M.tuberculosis_H37Rv AGG------------LFGVGGAGG--------------------------
MUL_0212|M.ulcerans_Agy99 AGGV-----------FFGNGGAGG--------------------------
MMAR_0607|M.marinum_M ATGI-----------FGGTGGAGG--------------------------
Mflv_5091|M.gilvum_PYR-GCK AG-----------DGTGGTGGTGG----TGGQGGQGGQGG----------
Mvan_1264|M.vanbaalenii_PYR-1 AG-----------DGLGGTGGTGG----TGGFGGQGGQGG----------
MSMEG_0868|M.smegmatis_MC2_155 AGRIRVNNGTATGTATGGNGGNGGNGDADGHRGGKGGDGGEALIRATVIG
TH_3810|M.thermoresistible__bu DG---------------GNGGSAG--------------------------
MAB_4284c|M.abscessus_ATCC_199 AG-------------LAGNAGLAG--------------------------
MLBr_01183|M.leprae_Br4923 --------------------------------------------------
MAV_1346|M.avium_104 --------------------------------------------------
Mb1849c|M.bovis_AF2122/97 -------AGGIG--------------------------------------
Rv1818c|M.tuberculosis_H37Rv -------AGGIG--------------------------------------
MUL_0212|M.ulcerans_Agy99 -------AGGTGGDGGGTGGAGGAA-----------------------GN
MMAR_0607|M.marinum_M -------AGGQSTN------------------------------------
Mflv_5091|M.gilvum_PYR-GCK -------TGGTGGQGG-LGGGGGTG-----------------------GQ
Mvan_1264|M.vanbaalenii_PYR-1 -------TGGTGGQGG-LGGVGGTG-----------------------GT
MSMEG_0868|M.smegmatis_MC2_155 SNASGTATGGDGGNGGTLGATNGTGGAATIVAYNVVQANPQASGTATDGH
TH_3810|M.thermoresistible__bu -------RGGTGGLRGVFGINGGNG------------------------R
MAB_4284c|M.abscessus_ATCC_199 -------NAAVAGNAAVAGNAAVAG------------------------N
MLBr_01183|M.leprae_Br4923 --------------------------------------------------
MAV_1346|M.avium_104 --------------------------------------------------
Mb1849c|M.bovis_AF2122/97 --LVGNGGAGGSGGSALLWGDG-----------------GAGGAGGVGST
Rv1818c|M.tuberculosis_H37Rv --LVGNGGAGGSGGSALLWGDG-----------------GAGGAGGVGST
MUL_0212|M.ulcerans_Agy99 GVLIGNGGNGGIGGAAGLWGTG-----------------GTGGSGGSGGT
MMAR_0607|M.marinum_M ALADSVGGNGGQGGDGRLFGNG-----------------GAGGVGGAAYT
Mflv_5091|M.gilvum_PYR-GCK GGTGGTGGQGGTGGTGGTGGTG--------GTGGAGGDGGLGGLGGFGGS
Mvan_1264|M.vanbaalenii_PYR-1 GGTGGTGGQGGTGGTGGTGGTG--------GTGGAGGDGGLGGLGGFGGV
MSMEG_0868|M.smegmatis_MC2_155 GGSGINGGHGGNGGQGTIEANGPSAVASGIGAGGNGGDGIDGGLGGNGGD
TH_3810|M.thermoresistible__bu AGLGGRGXXXXGGGVGAAVRSG---------------NGVFSGGGGTPSA
MAB_4284c|M.abscessus_ATCC_199 AAVGGVLNTGVSGAIGGSSHVG---------GAINESLGGSGSVGGSTAV
MLBr_01183|M.leprae_Br4923 --------------------------------------------------
MAV_1346|M.avium_104 --------------------------------------------------
Mb1849c|M.bovis_AF2122/97 ----------------TGGAGGAGG---------------------NAGL
Rv1818c|M.tuberculosis_H37Rv ----------------TGGAGGAGG---------------------NAGL
MUL_0212|M.ulcerans_Agy99 GATGGFSGLLASGDGGAGGAGGAGG---------------------AGGL
MMAR_0607|M.marinum_M ----------------------------------------------SNIL
Mflv_5091|M.gilvum_PYR-GCK GGTGG-QGGLGGPGGSTGGAGGAGG--QAGTAGKGGLFG----GSGSAGK
Mvan_1264|M.vanbaalenii_PYR-1 GGTGG-QGGQGGPGGSTGGAGGAGG--QAGVAGKGGLFG----GSGSAGT
MSMEG_0868|M.smegmatis_MC2_155 GVVNT-TANAANPGSATGTATGGDGGKATGTASQGGNGGPSWVQSGYGST
TH_3810|M.thermoresistible__bu GFGVP-TTIQFPAGSCTSRAVSREG------------------------I
MAB_4284c|M.abscessus_ATCC_199 GGGAH-TGVGGGVSTGVGVGSHVGG--------------------GISES
MLBr_01183|M.leprae_Br4923 --------------------------------------------------
MAV_1346|M.avium_104 --------------------------------------------------
Mb1849c|M.bovis_AF2122/97 L---VGAGG------------------------------AGGAGALGGGA
Rv1818c|M.tuberculosis_H37Rv L---VGAGG------------------------------AGGAGALGGGA
MUL_0212|M.ulcerans_Agy99 LDGLVGAGGGDGGAGGNAGTLFG----------------SGGVGGDGGSG
MMAR_0607|M.marinum_M MN---ATGG------------------------------NGGQGGTGGTL
Mflv_5091|M.gilvum_PYR-GCK AGATGAAGASGGKGAQGATGAQGATG------AQGATG-ATGAAGKAGAT
Mvan_1264|M.vanbaalenii_PYR-1 AGVTGASGASGGKGAQGATGAQGATG------AQGATG-ATGASGAQGSA
MSMEG_0868|M.smegmatis_MC2_155 LGGNATGTATAGKGGEASNGGVAGNGGNALVMAAGTTGVATGNTATGGDG
TH_3810|M.thermoresistible__bu MPSSRAASARAG---------------------------AACASSTSRSR
MAB_4284c|M.abscessus_ATCC_199 LGGSGSVGGSTAVGGGAHTGVGGGVS----------TGVGVGSHVGGGIS
MLBr_01183|M.leprae_Br4923 --------------------------------------------------
MAV_1346|M.avium_104 --------------------------------------------------
Mb1849c|M.bovis_AF2122/97 TGVGGAGGNGGTAGLL--------FGAGGAGGAGGFGFGGAGGAGGLGGK
Rv1818c|M.tuberculosis_H37Rv TGVGGAGGNGGTAGLL--------FGAG---GAGGFGFGGAGGAGGLGGK
MUL_0212|M.ulcerans_Agy99 HGNGGDGGGGGNAGLL--------FSDAGGGGFGGFGLAGGGGTGGSGGD
MMAR_0607|M.marinum_M FGNGGGGGTGG-------------------AGFVGPSSAGDGGNGGGGGR
Mflv_5091|M.gilvum_PYR-GCK GAAGAAGAAGSAG-----------TTGAQGSAGTNGA-NGANGSQGAAGK
Mvan_1264|M.vanbaalenii_PYR-1 GSAGSAGVAGSAG-----------TTGAQGSAGTNGA-NGANGSQGAAGK
MSMEG_0868|M.smegmatis_MC2_155 GAATTAGATGGAGGFSNIAAIHGAITDSTAISGHGGAGNGGNGGKGADGW
TH_3810|M.thermoresistible__bu RSFCCSRA------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 ESLGGSGSVGGSTAVG---------GGAHTGVGGGVNTGVGVGSHVGGGI
MLBr_01183|M.leprae_Br4923 --------------------------------------------------
MAV_1346|M.avium_104 --------------------------------------------------
Mb1849c|M.bovis_AF2122/97 AGLIGDG---------------------------GDGGAGGNG-------
Rv1818c|M.tuberculosis_H37Rv AGLIGDG---------------------------GDGGAGGNG-------
MUL_0212|M.ulcerans_Agy99 AGWLGSGGAGGAGGISVNGDAGAGGAGGTSGQLLGDGGAGGAGGEA----
MMAR_0607|M.marinum_M AGLIGNG---------------------------GAGGAGGAPGP-----
Mflv_5091|M.gilvum_PYR-GCK AGATGTTGATG--------AAGEDGDAGSGGSG-INGGAGGAGG------
Mvan_1264|M.vanbaalenii_PYR-1 AGATGTTGATG--------AAGDAGDPGSEAVD-INGGAGGAGG------
MSMEG_0868|M.smegmatis_MC2_155 VQAYDPAGPVGSKIANSSAVAGNGGDGGVNGTGGASGGDGGNGGNTSVWK
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 SESLGGSGSVG-------GSTAVGGGAHTSAINDALGGSGSVGG------
MLBr_01183|M.leprae_Br4923 --------------------------------------------------
MAV_1346|M.avium_104 --------------------------------------------------
Mb1849c|M.bovis_AF2122/97 --------TGAKGGDGGAGGGAILVGNGGNGGNAGSGTPNGS--------
Rv1818c|M.tuberculosis_H37Rv --------TGAKGGDGGAGGGAILVGNGGNGGNAGSGTPNGS--------
MUL_0212|M.ulcerans_Agy99 -----ELAAGGAGGSGGVGGDAVLIGAGGNGGNGGPGTAKGD--------
MMAR_0607|M.marinum_M -------NGGFSGGNGGNGGDAVLIGNGGNSGDVG-LSGAGT--------
Mflv_5091|M.gilvum_PYR-GCK -----AGGEGGEGGDGGKGGAGAEGGAGGGGGAGGSGAGGAG-------G
Mvan_1264|M.vanbaalenii_PYR-1 -----AGGEGGEGGAGGAGGDGSEGGAGGSGGAGGSGTGGAG-------G
MSMEG_0868|M.smegmatis_MC2_155 SGGQTADGEHNTGGNGGNGGDGSEPGPGGNGAGGAGGNGGTGGTTAGGSG
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 -STAVGGGAHTSAINESMNSSINASHSSSMNESFGSSMGSHGAAGTGFEA
MLBr_01183|M.leprae_Br4923 --------------------------------------------------
MAV_1346|M.avium_104 --------------------------------------------------
Mb1849c|M.bovis_AF2122/97 -AGTGGAGGLLGKNGMNGLP--------
Rv1818c|M.tuberculosis_H37Rv -AGTGGAGGLLGKNGMNGLP--------
MUL_0212|M.ulcerans_Agy99 -PGSGGAGGLLGVDGLNGLS--------
MMAR_0607|M.marinum_M -PGLPGNGGLL--IGTIGNT--------
Mflv_5091|M.gilvum_PYR-GCK AGGSGGSGGSGGSGGSGGAAGDSDD---
Mvan_1264|M.vanbaalenii_PYR-1 AGGSGGSGGSGGAGGSGGAAGDSD----
MSMEG_0868|M.smegmatis_MC2_155 NGGNGGNGGNGGADGTPGTAGTTGANNP
TH_3810|M.thermoresistible__bu ----------------------------
MAB_4284c|M.abscessus_ATCC_199 QGHAQGHASETAHTTLPGAEHH------
MLBr_01183|M.leprae_Br4923 ----------------------------
MAV_1346|M.avium_104 ----------------------------