For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NTSEYQVAGMRCGHCESAIRSEVAQIPGVATVAVSARTGRLLVAGSAPIDPTTVLGAIDEAGYQAVLVA*
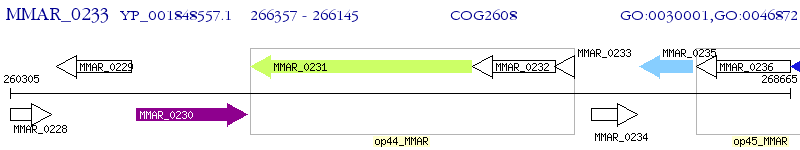
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0233 | - | - | 100% (70) | hypothetical protein MMAR_0233 |
| M. marinum M | MMAR_0231 | ctpA_1 | 6e-05 | 30.88% (68) | cation transporter p-type ATPase CtpA_1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2847 | - | 4e-16 | 52.17% (69) | heavy metal transport/detoxification protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0454c | - | 9e-12 | 45.31% (64) | putative metal-binding protein |
| M. avium 104 | MAV_5245 | - | 2e-23 | 70.00% (70) | hypothetical protein MAV_5245 |
| M. smegmatis MC2 155 | MSMEG_5016 | - | 3e-11 | 42.19% (64) | hypothetical protein MSMEG_5016 |
| M. thermoresistible (build 8) | TH_2912 | - | 1e-07 | 45.00% (60) | PUTATIVE Heavy metal transport/detoxification protein |
| M. ulcerans Agy99 | MUL_4881 | - | 8e-35 | 100.00% (70) | hypothetical protein MUL_4881 |
| M. vanbaalenii PYR-1 | Mvan_4448 | - | 6e-09 | 38.60% (57) | heavy metal transport/detoxification protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0233|M.marinum_M ---------------------------------------------MNTSE
MUL_4881|M.ulcerans_Agy99 ---------------------------------------------MNTSE
MAV_5245|M.avium_104 ---------------------------------------------MPTSE
Mflv_2847|M.gilvum_PYR-GCK MPRPASTRHQQEPEEPTLAPYPRGVYLDVRIPGRGTTESEDGSSVMSTIT
MSMEG_5016|M.smegmatis_MC2_155 ---------------------------------------------MTTQT
Mvan_4448|M.vanbaalenii_PYR-1 ---------------------------------------------MST-T
MAB_0454c|M.abscessus_ATCC_199 ---------------------------------------------MSTTA
TH_2912|M.thermoresistible__bu ----------------------------------------------MSVE
:
MMAR_0233|M.marinum_M YQVAGMRCGHCESAIRSEVAQIPGVATVAVSARTGRLLVAGSAPIDPTTV
MUL_4881|M.ulcerans_Agy99 YQVAGMRCGHCESAIRSEVAQIPGVATVAVSARTGRLLVAGSAPIDPTTV
MAV_5245|M.avium_104 YQVSGMSCGHCEAAVHSEVARIPGVDGVSVSADTGRLVVTSAVPIDADAV
Mflv_2847|M.gilvum_PYR-GCK YAVTGMTCGHCELSVREEVSGVAGVQDVEVSATAGTLIVTSSGPVDDAQI
MSMEG_5016|M.smegmatis_MC2_155 VTVTGMTCGHCVTSVREEVGSIPGVTTVDVDLATGQVTIGSDRQLDDEAI
Mvan_4448|M.vanbaalenii_PYR-1 ITVEGMSCAGCANSVRAELTHIPGVREVDVDISNGTVTIDSDNPVDPAAI
MAB_0454c|M.abscessus_ATCC_199 LKVSGMTCGHCVAAVREQVAAVPGVTGVQVDVATGTVTVTSSGPLPAEAV
TH_2912|M.thermoresistible__bu IHVRGMSCGHCVSAITSAVRPLPGVEGVEVDLDAGVVRVTGTP--DEGAV
* ** *. * :: : :.** * *. * : : . :
MMAR_0233|M.marinum_M LGAIDEAGYQAVLVA
MUL_4881|M.ulcerans_Agy99 LGAIDEAGYQAVLVA
MAV_5245|M.avium_104 LGAVDEAGFQAVLVA
Mflv_2847|M.gilvum_PYR-GCK LRAVDEAGYSAVRVV
MSMEG_5016|M.smegmatis_MC2_155 REAVAEAGYAVAS--
Mvan_4448|M.vanbaalenii_PYR-1 REAVEEAGYTLAG--
MAB_0454c|M.abscessus_ATCC_199 HAAIRAAGYALAS--
TH_2912|M.thermoresistible__bu TAAIEDCGYDVPGA-
*: .*: