For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPRPASTRHQQEPEEPTLAPYPRGVYLDVRIPGRGTTESEDGSSVMSTITYAVTGMTCGHCELSVREEVS GVAGVQDVEVSATAGTLIVTSSGPVDDAQILRAVDEAGYSAVRVV
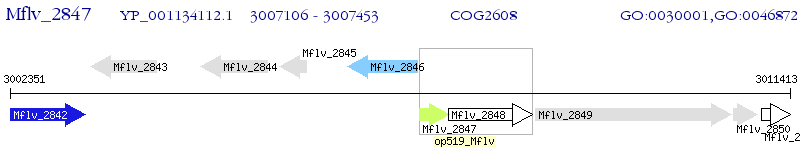
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2847 | - | - | 100% (115) | heavy metal transport/detoxification protein |
| M. gilvum PYR-GCK | Mflv_0910 | - | 3e-31 | 94.20% (69) | heavy metal transport/detoxification protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0454c | - | 3e-13 | 49.23% (65) | putative metal-binding protein |
| M. marinum M | MMAR_0233 | - | 5e-16 | 52.17% (69) | hypothetical protein MMAR_0233 |
| M. avium 104 | MAV_5245 | - | 8e-16 | 53.62% (69) | hypothetical protein MAV_5245 |
| M. smegmatis MC2 155 | MSMEG_5016 | - | 6e-13 | 50.77% (65) | hypothetical protein MSMEG_5016 |
| M. thermoresistible (build 8) | TH_2912 | - | 7e-07 | 38.60% (57) | PUTATIVE Heavy metal transport/detoxification protein |
| M. ulcerans Agy99 | MUL_4881 | - | 4e-16 | 52.17% (69) | hypothetical protein MUL_4881 |
| M. vanbaalenii PYR-1 | Mvan_3669 | - | 3e-11 | 46.38% (69) | heavy metal transport/detoxification protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0233|M.marinum_M ---------------------------------------------MNTSE
MUL_4881|M.ulcerans_Agy99 ---------------------------------------------MNTSE
MAV_5245|M.avium_104 ---------------------------------------------MPTSE
Mflv_2847|M.gilvum_PYR-GCK MPRPASTRHQQEPEEPTLAPYPRGVYLDVRIPGRGTTESEDGSSVMSTIT
MAB_0454c|M.abscessus_ATCC_199 ---------------------------------------------MSTTA
MSMEG_5016|M.smegmatis_MC2_155 ---------------------------------------------MTTQT
Mvan_3669|M.vanbaalenii_PYR-1 ---------------------------------------------MSTTT
TH_2912|M.thermoresistible__bu ---------------------------------------------MSVEI
* .
MMAR_0233|M.marinum_M YQ----VAGMRCGHCESAIRSEVAQIPGVATVAVSARTG---RLLVAGSA
MUL_4881|M.ulcerans_Agy99 YQ----VAGMRCGHCESAIRSEVAQIPGVATVAVSARTG---RLLVAGSA
MAV_5245|M.avium_104 YQ----VSGMSCGHCEAAVHSEVARIPGVDGVSVSADTG---RLVVTSAV
Mflv_2847|M.gilvum_PYR-GCK YA----VTGMTCGHCELSVREEVSGVAGVQDVEVSATAG---TLIVTSSG
MAB_0454c|M.abscessus_ATCC_199 LK----VSGMTCGHCVAAVREQVAAVPGVTGVQVDVATG---TVTVTSSG
MSMEG_5016|M.smegmatis_MC2_155 VT----VTGMTCGHCVTSVREEVGSIPGVTTVDVDLATG---QVTIGSDR
Mvan_3669|M.vanbaalenii_PYR-1 TTKSYAVTGMTCSHCIAAVSAELGALTGVTDVHVDLVAGGISTVTVSSDT
TH_2912|M.thermoresistible__bu H-----VRGMSCGHCVSAITSAVRPLPGVEGVEVDLDAG-----VVRVTG
* ** *.** :: : :.** * *. :* :
MMAR_0233|M.marinum_M PIDPTTVLGAIDEAGYQAVLVA---
MUL_4881|M.ulcerans_Agy99 PIDPTTVLGAIDEAGYQAVLVA---
MAV_5245|M.avium_104 PIDADAVLGAVDEAGFQAVLVA---
Mflv_2847|M.gilvum_PYR-GCK PVDDAQILRAVDEAGYSAVRVV---
MAB_0454c|M.abscessus_ATCC_199 PLPAEAVHAAIRAAG-YALAS----
MSMEG_5016|M.smegmatis_MC2_155 QLDDEAIREAVAEAG-YAVAS----
Mvan_3669|M.vanbaalenii_PYR-1 PLTVAQVATALDEAGDYRLSTDQPR
TH_2912|M.thermoresistible__bu TPDEGAVTAAIEDCG-YDVPGA---
: *: .* :