For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPTSEYQVSGMSCGHCEAAVHSEVARIPGVDGVSVSADTGRLVVTSAVPIDADAVLGAVDEAGFQAVLVA
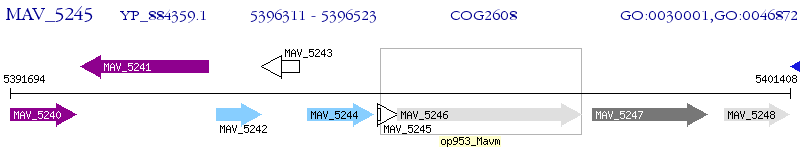
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_5245 | - | - | 100% (70) | hypothetical protein MAV_5245 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0106c | ctpB | 4e-06 | 42.37% (59) | cation-transporter P-type ATPase B |
| M. gilvum PYR-GCK | Mflv_0910 | - | 5e-16 | 54.29% (70) | heavy metal transport/detoxification protein |
| M. tuberculosis H37Rv | Rv0092 | ctpA | 2e-05 | 35.94% (64) | cation transporter P-type ATPase A |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0454c | - | 4e-14 | 51.56% (64) | putative metal-binding protein |
| M. marinum M | MMAR_0233 | - | 2e-23 | 70.00% (70) | hypothetical protein MMAR_0233 |
| M. smegmatis MC2 155 | MSMEG_5016 | - | 4e-10 | 43.75% (64) | hypothetical protein MSMEG_5016 |
| M. thermoresistible (build 8) | TH_2912 | - | 2e-09 | 45.00% (60) | PUTATIVE Heavy metal transport/detoxification protein |
| M. ulcerans Agy99 | MUL_4881 | - | 1e-23 | 70.00% (70) | hypothetical protein MUL_4881 |
| M. vanbaalenii PYR-1 | Mvan_4448 | - | 3e-08 | 42.11% (57) | heavy metal transport/detoxification protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0233|M.marinum_M -------------MNTSEYQVAGMRCGHCESAIRSEVAQIPGVATVAVSA
MUL_4881|M.ulcerans_Agy99 -------------MNTSEYQVAGMRCGHCESAIRSEVAQIPGVATVAVSA
MAV_5245|M.avium_104 -------------MPTSEYQVSGMSCGHCEAAVHSEVARIPGVDGVSVSA
Mflv_0910|M.gilvum_PYR-GCK -------------MSTITYAVTGMTCGHCELSVREEVSGVAGVEDVEVSA
MSMEG_5016|M.smegmatis_MC2_155 -------------MTTQTVTVTGMTCGHCVTSVREEVGSIPGVTTVDVDL
Mvan_4448|M.vanbaalenii_PYR-1 -------------MST-TITVEGMSCAGCANSVRAELTHIPGVREVDVDI
MAB_0454c|M.abscessus_ATCC_199 -------------MSTTALKVSGMTCGHCVAAVREQVAAVPGVTGVQVDV
TH_2912|M.thermoresistible__bu -------------MSV-EIHVRGMSCGHCVSAITSAVRPLPGVEGVEVDL
Mb0106c|M.bovis_AF2122/97 MAAPVVGDADLQSVRRIRLDVSGMSCAACASRVETKLNKIPGVR-ASVNF
Rv0092|M.tuberculosis_H37Rv MTTAVTG-EHHASVQRIQLRISGMSCSACAHRVESTLNKLPGVR-AAVNF
: : ** *. * : : :.** . *.
MMAR_0233|M.marinum_M RTGRLLVAGSAPIDPTTVLGAIDEAGYQAVLVA-----------------
MUL_4881|M.ulcerans_Agy99 RTGRLLVAGSAPIDPTTVLGAIDEAGYQAVLVA-----------------
MAV_5245|M.avium_104 DTGRLVVTSAVPIDADAVLGAVDEAGFQAVLVA-----------------
Mflv_0910|M.gilvum_PYR-GCK TTGTLIVTSSGLVDDAQILHAVDEAGYSAVRVA-----------------
MSMEG_5016|M.smegmatis_MC2_155 ATGQVTIGSDRQLDDEAIREAVAEAGYAVAS-------------------
Mvan_4448|M.vanbaalenii_PYR-1 SNGTVTIDSDNPVDPAAIREAVEEAGYTLAG-------------------
MAB_0454c|M.abscessus_ATCC_199 ATGTVTVTSSGPLPAEAVHAAIRAAGYALAS-------------------
TH_2912|M.thermoresistible__bu DAGVVRVTGTP--DEGAVTAAIEDCGYDVPGA------------------
Mb0106c|M.bovis_AF2122/97 ATRVATID-AVGMAADELCGVVEKAGYHAAPHTETTVLDKRTKDPDGAHA
Rv0092|M.tuberculosis_H37Rv GTRVATIDTSEAVDAAALCQAVRRAGYQADLCTDD---GRSASDPDADHA
: : .: .*:
MMAR_0233|M.marinum_M --------------------------------------------------
MUL_4881|M.ulcerans_Agy99 --------------------------------------------------
MAV_5245|M.avium_104 --------------------------------------------------
Mflv_0910|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5016|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_4448|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0454c|M.abscessus_ATCC_199 --------------------------------------------------
TH_2912|M.thermoresistible__bu --------------------------------------------------
Mb0106c|M.bovis_AF2122/97 RRLLRRLLVAAVLFVPLADLSTLFAIVPSARVPGWGYILTALAAPVVTWA
Rv0092|M.tuberculosis_H37Rv RQLLIRLAIAAVLFVPVADLSVMFGVVPATRFTGWQWVLSALALPVVTWA
MMAR_0233|M.marinum_M --------------------------------------------------
MUL_4881|M.ulcerans_Agy99 --------------------------------------------------
MAV_5245|M.avium_104 --------------------------------------------------
Mflv_0910|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5016|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_4448|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0454c|M.abscessus_ATCC_199 --------------------------------------------------
TH_2912|M.thermoresistible__bu --------------------------------------------------
Mb0106c|M.bovis_AF2122/97 AWPFHSVALRNARHRTTSMETLISVGIVAATAWSLSSVFGDQPPREGSGI
Rv0092|M.tuberculosis_H37Rv AWPFHRVAMRNARHHAASMETLISVGITAATIWSLYTVFGNHSPIERSGI
MMAR_0233|M.marinum_M --------------------------------------------------
MUL_4881|M.ulcerans_Agy99 --------------------------------------------------
MAV_5245|M.avium_104 --------------------------------------------------
Mflv_0910|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5016|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_4448|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0454c|M.abscessus_ATCC_199 --------------------------------------------------
TH_2912|M.thermoresistible__bu --------------------------------------------------
Mb0106c|M.bovis_AF2122/97 WRAILNSDSIYLEVAAGVTVFVLAGRYFEARAKSKAGSALRALAELGAKN
Rv0092|M.tuberculosis_H37Rv WQALLGSDAIYFEVAAGVTVFVLVGRYFEARAKSQAGSALRALAALSAKE
MMAR_0233|M.marinum_M --------------------------------------------------
MUL_4881|M.ulcerans_Agy99 --------------------------------------------------
MAV_5245|M.avium_104 --------------------------------------------------
Mflv_0910|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5016|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_4448|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0454c|M.abscessus_ATCC_199 --------------------------------------------------
TH_2912|M.thermoresistible__bu --------------------------------------------------
Mb0106c|M.bovis_AF2122/97 VAVLLPDGAELVIPASELKKRQRFVTRPGETIAADGVVVDGSAAIDMSAM
Rv0092|M.tuberculosis_H37Rv VAVLLPDGSEMVIPADELKEQQRFVVRPGQIVAADGLAVDGSAAVDMSAM
MMAR_0233|M.marinum_M --------------------------------------------------
MUL_4881|M.ulcerans_Agy99 --------------------------------------------------
MAV_5245|M.avium_104 --------------------------------------------------
Mflv_0910|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5016|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_4448|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0454c|M.abscessus_ATCC_199 --------------------------------------------------
TH_2912|M.thermoresistible__bu --------------------------------------------------
Mb0106c|M.bovis_AF2122/97 TGEAKPVRAYPAASVVGGTVVMDGRLVIEATAVGADTQFAAMVRLVEQAQ
Rv0092|M.tuberculosis_H37Rv TGEAKPTRVRPGGQVIGGTTVLDGRLIVEAAAVGADTQFAGMVRLVEQAQ
MMAR_0233|M.marinum_M --------------------------------------------------
MUL_4881|M.ulcerans_Agy99 --------------------------------------------------
MAV_5245|M.avium_104 --------------------------------------------------
Mflv_0910|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5016|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_4448|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0454c|M.abscessus_ATCC_199 --------------------------------------------------
TH_2912|M.thermoresistible__bu --------------------------------------------------
Mb0106c|M.bovis_AF2122/97 TQKARAQRLADHIAGVFVPVVFVIAGLAGAAWLVSGAGADRAFSVTLGVL
Rv0092|M.tuberculosis_H37Rv AQKADAQRLADRISSVFVPAVLVIAALTAAGWLIAGGQPDRAVSAALAVL
MMAR_0233|M.marinum_M --------------------------------------------------
MUL_4881|M.ulcerans_Agy99 --------------------------------------------------
MAV_5245|M.avium_104 --------------------------------------------------
Mflv_0910|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5016|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_4448|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0454c|M.abscessus_ATCC_199 --------------------------------------------------
TH_2912|M.thermoresistible__bu --------------------------------------------------
Mb0106c|M.bovis_AF2122/97 VIACPCALGLATPTAMMVASGRGAQLGIFIKGYRALETIRSIDTVVFDKT
Rv0092|M.tuberculosis_H37Rv VIACPCALGLATPTAMMVASGRGAQLGIFLKGYKSLEATRAVDTVVFDKT
MMAR_0233|M.marinum_M --------------------------------------------------
MUL_4881|M.ulcerans_Agy99 --------------------------------------------------
MAV_5245|M.avium_104 --------------------------------------------------
Mflv_0910|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5016|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_4448|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0454c|M.abscessus_ATCC_199 --------------------------------------------------
TH_2912|M.thermoresistible__bu --------------------------------------------------
Mb0106c|M.bovis_AF2122/97 GTLTVGQLAVSTVTMAGSGTSERDREEVLGLAAAVESASEHAMAAAIVAA
Rv0092|M.tuberculosis_H37Rv GTLTTGRLQVSAVTAAPG----WEADQVLALAATVEAASEHSVALAIAAA
MMAR_0233|M.marinum_M --------------------------------------------------
MUL_4881|M.ulcerans_Agy99 --------------------------------------------------
MAV_5245|M.avium_104 --------------------------------------------------
Mflv_0910|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5016|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_4448|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0454c|M.abscessus_ATCC_199 --------------------------------------------------
TH_2912|M.thermoresistible__bu --------------------------------------------------
Mb0106c|M.bovis_AF2122/97 SPDPGPVNGFVAVAGCGVSGEVGGHHVEVGKPSWITRTTPCHDAALVSAR
Rv0092|M.tuberculosis_H37Rv TTRRDAVTDFRAIPGRGVSGTVSGRAVRVGKPSWIG-SSSCHPN-MRAAR
MMAR_0233|M.marinum_M --------------------------------------------------
MUL_4881|M.ulcerans_Agy99 --------------------------------------------------
MAV_5245|M.avium_104 --------------------------------------------------
Mflv_0910|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5016|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_4448|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0454c|M.abscessus_ATCC_199 --------------------------------------------------
TH_2912|M.thermoresistible__bu --------------------------------------------------
Mb0106c|M.bovis_AF2122/97 LDGESRGETVVFVSVDGVVRAALTIADTLKDSAAAAVAALRSRGLRTILL
Rv0092|M.tuberculosis_H37Rv RHAESLGETAVFVEVDGEPCGVIAVADAVKDSARDAVAALADRGLRTMLL
MMAR_0233|M.marinum_M --------------------------------------------------
MUL_4881|M.ulcerans_Agy99 --------------------------------------------------
MAV_5245|M.avium_104 --------------------------------------------------
Mflv_0910|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5016|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_4448|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0454c|M.abscessus_ATCC_199 --------------------------------------------------
TH_2912|M.thermoresistible__bu --------------------------------------------------
Mb0106c|M.bovis_AF2122/97 TGDNRAAADAVAAQVGIDSAVADMLPEGKVDVIQRLREEGHTVAMVGDGI
Rv0092|M.tuberculosis_H37Rv TGDNPESAAAVATRVGIDEVIADILPEGKVDVIEQLRDRGHVVAMVGDGI
MMAR_0233|M.marinum_M --------------------------------------------------
MUL_4881|M.ulcerans_Agy99 --------------------------------------------------
MAV_5245|M.avium_104 --------------------------------------------------
Mflv_0910|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5016|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_4448|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0454c|M.abscessus_ATCC_199 --------------------------------------------------
TH_2912|M.thermoresistible__bu --------------------------------------------------
Mb0106c|M.bovis_AF2122/97 NDGPALVGADLGLAIGRGTDVALGAADIILVRDDLNTVPQALDLARATMR
Rv0092|M.tuberculosis_H37Rv NDGPALARADLGMAIGRGTDVAIGAADIILVRDHLDVVPLALDLARATMR
MMAR_0233|M.marinum_M --------------------------------------------------
MUL_4881|M.ulcerans_Agy99 --------------------------------------------------
MAV_5245|M.avium_104 --------------------------------------------------
Mflv_0910|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5016|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_4448|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0454c|M.abscessus_ATCC_199 --------------------------------------------------
TH_2912|M.thermoresistible__bu --------------------------------------------------
Mb0106c|M.bovis_AF2122/97 TIRMNMIWAFGYNVAAIPIAAAGLLNPLIAGAAMAFSSFFVVSNSLRLRN
Rv0092|M.tuberculosis_H37Rv TVKLNMVWAFGYNIAAIPVAAAGLLNPLVAGAAMAFSSFFVVSNSLRLRK
MMAR_0233|M.marinum_M ----------------------
MUL_4881|M.ulcerans_Agy99 ----------------------
MAV_5245|M.avium_104 ----------------------
Mflv_0910|M.gilvum_PYR-GCK ----------------------
MSMEG_5016|M.smegmatis_MC2_155 ----------------------
Mvan_4448|M.vanbaalenii_PYR-1 ----------------------
MAB_0454c|M.abscessus_ATCC_199 ----------------------
TH_2912|M.thermoresistible__bu ----------------------
Mb0106c|M.bovis_AF2122/97 FGAQ------------------
Rv0092|M.tuberculosis_H37Rv FGRYPLGCGTVGGPQMTAPSSA